Current Equipment
Ross Fluorescence Imaging Center Equipment Summary, including ACCM Leica SP8 confocal microscope (we manage for ACCM). Fluorophore tips at bottom of first table.
Image core manager George McNamara, PhD. gmcnamara2jhmi.edu
August 2024 iLab URL change - my JH link should update but you may need to manually fix your bookmarks (note - the iLab links in the text table below have not been updated):
https://johnshopkins.ilab-int.agilent.com
Full notice: Please be aware that as of 8/3/2024 (late Friday night) the URL for the Johns Hopkins University iLab website will be changing to https://johnshopkins.ilab-int.agilent.com. Any previously saved bookmarks for iLab should be updated accordingly. If you encounter any issues with logging into iLab with your JHED ID, please call the Johns Hopkins IT Help Desk at 410-516-HELP (410-516-4357) or submit a Johns Hopkins Help Desk self-service ticket online by accessing https://myit.jh.edu/apps/mfahelpticket.
Reservations (iLab URLs changed in 2024)
Ross image core https://johnshopkins.ilab.agilent.com/sc/5148/ross-imaging-center/?tab=equipment
ACCM LeicaSP8 confocal microscope https://johnshopkins.ilab.agilent.com/schedules/345585#/schedule
* as of 7/2023 George is 20% time in Ross bldg, 80% time in Smith Bldg managing the High Throughput Phenotypic Screening Core (HPS core) - please plan ahead for anything to do with the Ross Imaging Core or ACCM Leica SP8 confocal microscope core.
* Please note Olympus is now known as Evident Scientific.
|
August 2024 tip: We are not the only image core at JHU. You may find unique instruments/services and/or quicker responsive staff (I am now 20% time in Ross; 80% time elsewhere),for examples: * Aleksandr V. Smirnov, PHD & MPSasmirno3@jhmi.edu,Neuroscience Imaging Center (SOM, PCTB) has an Abberior Facility Line STED microscope (nanoscope), 775nm depletion laser line (i.e. Abberior STAR RED). * Ian Dobbie, PhD, IIC Homewood campus, has lots of fluorescence microscopes, electron microscopes, sample preparation. * eye research - Wilmer Eye Institute (Smith Bldg) has an imaging core, primarily for vision / eye researchers. * JHU SOM has a Titan Krios (state of the art) cryo-EM (the space renovation may have cost more than your house). |
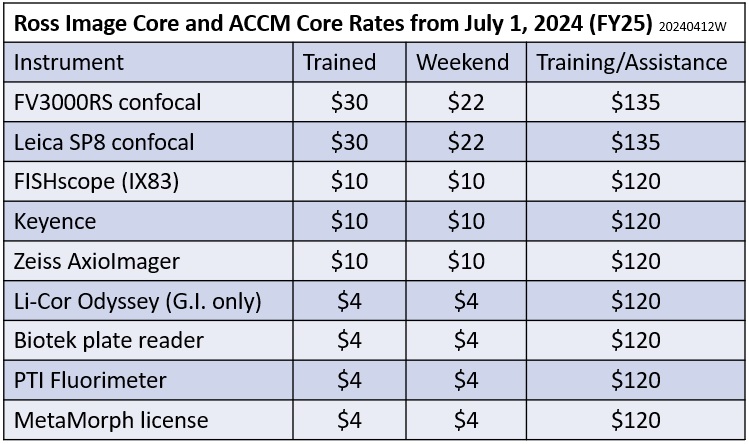
Rates as of July 1, 2025 (JHU FY25) shown below.
Covid-19 pandemic vaccine, mask, social distancing precautions - also flu vaccine please
September 14, 2022: JHU SOM In nonclinical buildings: As of Thursday, Sept. 15, masks are recommended but no longer required for JHM personnel working in nonclinical buildings where there is NO reasonable expectation of an encounter with a patient.
Ross Image Core policy - Sept 14, 2022: mask is optional for FULLY VACCINATED users -- as up to date covid=19 vaccines, boosters, new version(s) -- original+Onicron bivalent mRNA vaccines just came out. If you are not fully vaccinated, please wear a mask in (and near) the image core -- or arrange for a fully vaccinated colleague to do your imaging. If you do wear a mask, please wear it correctly.
Please continue to social distance as much as practical.
I note that covid-19 still kills AND long covid-19 sucks, seer
https://www.theatlantic.com/health/archive/2022/09/long-covid-brain-fog-symptom-executive-function/671393/
https://www.theatlantic.com/newsletters/archive/2022/09/long-covid-misunderstood-symptom-brain-fog/671435/
FLU Season precautions - pleasse get your influenza 2022 vaccine "as soon as practical". JHU SOM deadline is mid-November 2022. Available now (Sept 15, 2022) - GM walked over to campus Walgreens, got both the Pfizer bivalent covid-19 vaccine (one arm) and the influenza vaccine (other arm, paid $66 for 'instant gratification' instead of waiting to visit my health care provider or waiting for JHU SOM to offer the flu shot on campus).
|
Aug 15, 2022: recording session screens and audio - Leica SP8, Olympus FV3000RS confocal microscope PCs and George's PCs are now equipped with NCD Debut Pro recording software. Recordings should be "JHU internal use only" not posted on youtube etc.because the microscopy companies may have copyrights on their software user interface. Also good to speak slowly, move mouse cursor slowly and viewer may not be able to see all the action (etc no web cam, or bad view --> little or no 'action' at the microscope). We are open to adding this or similar software on other microscope PCs (NCH Debut Pro is typically $39.99 per PC). GM recommends practicing and making sure audio microphone - and if present web cam - are workign BEFORE starting a "production"; video (ex. a few days before AND 30 min before start). Recording problems can often be sorted out quickly (if you know Windows). GM also notes that Zoom has the ability to record a Zoom session (usually only the Host can do that). Aug 9, 2022 iLab issues for end users and Core Managers, JHU has a full set of simplified help documents on our Core in the Box Website. See link below. The available help docs on this page may not always solve the issue, but please feel free to send the link along to your users that may have questions. https://www.hopkinsmedicine.org/research/resources/synergy/core-in-a-box/ilab-support.html Also, emails for JHU contacts are: gmcnamara@jhmi.edu (me - for cores I am involved in), Shawn Franckowiak sfrancko@jhmi.edu , Jeffrey Smith smithje@jhmi.edu , Melanie Branagan mbranag1@jhmi.edu |
New (2021) service: single molecule FISH probe set generation, specimen labling optimization, training users to label your own specimens. See FISH reagents iLab web page.
|
Reminder: All users need to make reservations in iLab for whatever instrument/service they are using. We need this for both billing and to track usage for our NIH P30 Center grant and other administrative purposes. * upload your data to your JHU Microsoft OneDrive (myJH -> Cloud --> JH OneDrive [click heart button to make a favorite). no USB sticks allowed (subject to confisacation and discard in biosafety trash). Limit your web surfing to web sites/pages of JHU and critical vendors (not your personal web emasil etc) and especially no surfing the 'dark web' on image core computers. Non-JHU users: George can make reservations for you, and iLab can bill you through a credit card (iLab sends to JHU's SAP financial system, which then handles the credit card stuff). 20241206F: GM deleted ilab links at right because the old ones became defunct August 2024. Please go to the two main pages below: Ross Image Core https://johnshopkins.ilab.agilent.com/sc/5148/ross-imaging-center/?tab=equipment ACCM Confocal Microscope Core (Leica SP8 and Incucyte S3 -- latter is ACCM use only) https://johnshopkins.ilab.agilent.com/service_center/3804/?tab=equipment |
Information click on entry in this column or scroll down this page. |
iLab Organizer see links at left |
Workday hourly rate JHU trained user |
|
|
Olympus FV3000RS confocal ==> Please note Olympus is now known as Evident Scientific. |
Olympus FV3000RS info | $30 / hr | ||
|
Leica SP8 confocal (ACCM confocal core) 20240809F Leica SP8 HyD detectors linear range up to 60 photons per second (each) - see http://confocal.jhu.edu/mctips/leica-sp8-hyds-linear-range-to-60-photons-per-microsecond
|
Leica SP8 info | $30 / hr | ||
|
IX83 FISHscope ... widefield single molecule RNA fluorescence in situ hybridization (FISH), immunofluorescence Olympus cellSens Dimension 3.x info (advanced features) quick tips http://confocal.jhu.edu/mctips/fishscope-quick-tips including pricing estimates for several products ==> Please note Olympus is now known as Evident Scientific. |
$10 / hr | |||
|
Keyence BZ-X710 ... SBS plate, 35mm or 50mm dish (plastic or imaging dish work), microscope slide. We also have a four slide holder, which may let you scan up to four slides in one click (per channel). Note: data sets can get "pretty big"so if your officePC is slow, you may not be able to do much with your data (we have suggestions for improving PC performance on a budget). * objective lenses are all "low mag, modest NA, relatively long working distance" 4x, 10x, 20x. If you need high resolution, have specimen on coverglass (not SBS plate or tissue culture thick plastic) and learn our Leica SP8 and/or Olympus FV3000RS confocal microscopes or FISHscope widefield microscope(s). You are welcome to bring your own Nikon objective lens(es) and either use for session or loan to the image core -- if you loan a lens and it gets damaged by a user, we will not reimburse you and you should assume JHU will not force the user or their PI to reimburse you. * keyence works nicely for fluorescent slide scanning. For speed, acquire one channel at a time (ex. CH1, scan, CH2, scan, CH3, scan, CH4, scan). You can staet stitching (optional shading correction with 'white reference" image) and then alt-tab back to acquisiton software, change channel, acquire.
|
Keyence BZ-X710 info | $10 / hr | ||
|
Zeiss AxioObserver.A1 inverted microscope & Olympus DP80 dual RGB and monochrome camera ==> Please note Olympus is now known as Evident Scientific. |
Zeiss AxioImager info | $10 / hr | ||
| Li-Cor Odyssey | Li-Cor Odyssey CLx info | $4 / hr | ||
|
Biotek absorbance plate reader ... has five filters: 405, 450, 490, 630, 750nm Note: on 20220215U we replaced 415 with 750nm filter (6th position is "plug" = black). Model ELx808: For 96-well plates, 380 – 900 nm wavelength range and shaking BioTek/Agilent offers filters from 340nm to 750nm, so if someone would like to buy an alternative filters, we would be happy to replace one of the current filters(ex: 450nm --> 750nm). See "Filters" (drop down list) at https://www.biotek.com/products/accessories for list of filters available. |
(will add info page later - in meanwhile, see text to left) | $4 / hr | ||
|
PTI QuantaMaster 40 Fluorimeter #1 20241206F update: #1 in Ross S913 is currently buried under various things we moved from a former image core room. Do not assume you will be able to walk in and use this fluorimeter. |
PTI Fluorimeter #1 | $4 / hr | ||
|
PTI QuantaMaster 40 Fluorimeter #2 20241206F update: #2 has been dismantled and moved to a G.I. storage space. Several users of both #1 (see above) and #2 claimed that #1 gave better data (GM never saw any data for or against this). G.I. repurposed the room #2 was in (hopefully that space will generate lots of NIH grants and discoveries). |
||||
|
MetaMorph image analysis software 3 licenses (USB license keys can float) |
(#4 ex-Revolution X1 MM) |
|
$4 / hr $4 / hr $4 / hr ... |
|
|
See Usage Fees and Rates page for weekend, assisted and training pricing. Legacy equipment (retired) idescribed briefly at bottom of this web page https://johnshopkins.corefacilities.org/sc/5148/ross-imaging-center/?tab=services |
..................................... |
see left column | ||
|
April 2022 update: our NIH NIDDK funding ended and resubmissions were declined so we are putting "RNA FISH as a Service" on hold with respect to image core. Please contact Prof. Bin Wu directly (bwu20@jhmi.edu) about fesibility of you collaboratign with Bin and Wu lab on "going FISHing" together. Tip: background is a killer ... and there are a lot of sources of background when doing fluorescence. At some point, you should RNAse (maybe multiple RNAses) some test specimens as a negative control ... ideally your cells or tissues have low background, and RNAse(s) will make them a little lower. However, if your probe sets bind "non-specifically" to RNA, then RNAse(s) should lower background. Caveat: if you contaminate your lab with RNase(s) you'll never have a successful experiment in the future! Insufficient washes, and/or washes with the wrong solutions, or letting the specimen dry (even for a second), can all be bad. You may find useful testing some vendors wash reagents with our probe sets, i.e. RNAscope reagents and protocol (except for product specific stuff) or Biosearch Technologies "Stellaris FISH reagents (and some probe sets for comparison, i.e. RNA Pol IIA = PolR2A). |
||||
|
Usual fluorophores for immunofluorescence for our two confocal microscopes (Leica SP8, Olympus FV3000RS) widefield microscopes (FISHscope, Zeiss AxioObserver, Keyence): DAPI ... DNA counterstain Alexa Fluor 488 Alexa Fluor 568 gm note: Alexa Fluor 610-X is much brighter than AF594, Texas Red, ATTO 594 or Cy3.5 (Maillard 2020). Alexa Fluor 610-X is much brighter dye in solution, someone(s) will need to test on antibodies, oligos, etc. Alexa Fluor 647 Alexa Fluor 750 ... 5th fluorophore (best on FISHscope); Alexa Fluor 750, IRDye800, Cy7 (I note that NIR-conjugates may not be stable in moderate term storage -- NIR fluorophores tend to 'decompose' even in the dark - up to you to figure out proper storage and QA/QC that they still work ... they may fall aparet on antibodies or oligos). AF750 works well on FISHscope (widefield microscope, right light source, right Penta filter cube, right filters in emission filter wheel, right camera, excellent objective lenses). Good spectral viewer: https://searchlight.semrock.com/ a couple of reviews that may (or not) be useful: McNamara 2017 CPHG https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cphg.42 McNamara 2020 CTR - New Technologies (immunofl, smFISH, etc). McNamara 2014 MMB1075 - low mgnification confocal microscopy of tumor angiogenesis (aka "blood vessel painting - DiI works great) McNamara 2006 CSHL Imaging book - Introduction to immunofluorescence microscopy (JHU colleagues can get PDF from me). More multiplexing options -- starting with repplacing DAPI with BV421 and buildign on that with other modern fluorophores (if still need DNA counterstain could consider Zombie NIR): http://confocal.jhu.edu/mctips/multiplex * Consider modernizing your antibodies with direct labels (ex. BV421) or secondary nanobodies (ThermoFisher/Chromotek, Nano-Tag, Pleiner et al 2018 JCB published sequences in their supplemental file, so could generate in your own lab). A nice example showing advantage of nanobody over primary + seondary antibody is doi.org/10.1002/cphc.202100185 * I still like tyamide signal amplification (TSA), and am intrigued by rolling circle amplification (RCA) for "one shot" multiplexing" (see Ultivue patents). I am especially intrigued by DNA-PAINT for multiplexing (i.e. nanobody#1-oligo#1 detect with fluorescent complementary oligo#1) for single molecule localization (subsaturing c-oligo) or modernized immunofl (saturating c-oligo). -- Bad antibodies waste time and money. Two quotes: "Old soldiers never die, they just fade away." - Douglas Macarthur (long dead). :Old antibodies die, please throw them away." - GM. |
||||
|
VyeScan microscope slide scanner Pathscan Enabler like simple slide scanner (brightfield only) We have a 35 mm slide scanner and microscope slide scanner similar to the Pathscan Enabler V https://www.meyerinst.com/pathscan-enabler-5 (GM managed an original model back in 2000 at CHLA). This is a modest resolution scanner, "about equivalent resolution" to a typicaly 4x objective lens, but much bigger field of view (36x24 mm) and very fast scanning (~1 minute at ~4000 dpi, ~4 um pixel size). For an old example (Pathscan I) download Tiki_Goddess from https://works.bepress.com/gmcnamara/1 and zoom in to see details -- my office has a poster (printed on 'fabric') that is ~84x44 inches (84" = 7 feet, about the height of an NBA basketball center). I routinely use Hamrick Software VueScan Professional https://www.hamrick.com/purchase-vuescan.html ($99.95, license can go on a couple of computers) for slide scanner and flatbed scanner operation. VueScan Pro operates practically all scanners on the market (even obsolete scanners) and enables more control than commercial products -- and "knows" special features such as transparency lids. -->I also note that an office flatbed scanner can be used to scan in many brightfield (H&E, IHC, etc) slides in "one click" - so you could try whatever scanner is in your office or lab (and then try and buy VueScan Pro to make your scanning life better). Maximum resolution - max DPI - may not be needed (and may note be useful). For published examples with original Pathscan (circa 2000) see https://pubmed.ncbi.nlm.nih.gov/12451475 and https://pubmed.ncbi.nlm.nih.gov/12533523 Avramis ... McNamara ... Avramis et al 2002 In vitro and in vivo evaluations of the tyrosine kinase inhibitor NSC 680410 against human leukemia and glioblastoma cell lines. Cancer Chemother Pharmacol 50: 479-489. doi: 10.1007/s00280-002-0507-6.
|
in iLab | to be determined |
HPS Core - As of July 1, 2022, Dr. McNamara is also manager of High Throughput Phenotypic Screening Core (HPS Core) in Smith Bldg (Wilmer Eye Institute)
|
HPS Core enables large object phenotypic screening - typically of zebrafish embryos and larvae, C. elegans (worms), organoids, tumor spheroids. Key concept is to do initial High Troughput Screen (HTS) on a multi-mode plate reader, read out the data in real time, and transfer the candidate "hits" with a "large object sorter" (Union Biometrica COPAS / VAST) to imaging components. Prof. Jeff Mumm and collaborators have been doing this for over one decade and NIH high end instrumentation grant ($1.94M) was funded for the newest implementationm "ARQivST" (aka HPS2). https://grantome.com/grant/NIH/S10-OD026909-01A1 (also listed in NIH RePORTER .. at time of writign this text the search was not working). More details on HPS Core components on our iLab web page https://johnshopkins.ilab.agilent.com/sc/5771/high-throughput-phenotypic-screening-core/?tab=about Components summary * Hudson Robotics - plate crane and software to integrate all components - two Micro10x dispensers (minimum volume 10 uL per well); one with plate oriented each way, dispense with 8 tips or 12-tips (plate crane can position onto either). - SOLO dispense/aspiration pipetting station with 6 positions, i.e. 3 for plates, 3 for pipet tip racks. Typically one plate of larvae at at time, one plate with drugs (say 80 drugs, 12 DMSO only control wells), one plate for dilutions. - two plate cranes: SciSlopsSTD (standard), SciClopsRA (random access) -- reach every "platform" (plate position, i.e. Tecan Spark load plate). - 10 hotel carousel, each "hotel" can hold up to 30 plates or up to 7 pipet tip racks. - "Hopkins Handoff" - 6 position platform in the middle of the HPS to position a plate between the two plate cranes to "handoff" between them. * Tecan Spark multi-mode plate reader (state of the art ... bioluminescence, fluorescence, absorption ... best luciferase is Promega's NanoLuc, as in Jay Thierer & Steve Farber's LipoGlo zebrafish papers). Spark tip - top reads: do your best to make the meniscus a "flat top" for every well (same amount of liquid in each) -- convex or concave meniscus can act as a lens. * Union Biometrica 1. Biosorter ... large object sorter, for example 3000 zebrafish larvae (5 days post fertilization dimensions "diameter" ~600um, ~2 mm length) in 750 mL sample cup, 1000 um "FOCA" (flow cell), sort with excellent purity ~1 per second into 50 mL tube or one larva per well onto 96-well plate. If I have a lot of larvae, I like to "pre-sort" into 50 mL tube to get a single phenotype - such as "mid-brightness yellow fluorescent protein in rtinal ganglion cells", then put back in sample cup, and sort onto whatever number 96-well plates we need for screening. 2. LPSD-VAST - large particle sampler dispenser - vertebebrate automated screening technology * Applied Scientific Instrumentation ... LSFM light sheef fluorescence microscope - aka single sided SPIM (selective plane illumination microscope) - Goal: similar capability to Union's VASTomography (our JHU collaborators are working on the details) - ASI SPIM components, including 500um Piezo-Z drive, synchronized with the MEMS scanner light sheet, to acquire Z-series (optional: VAST can rotate capillary, then acquire more views). - Thorlabs Water Dipping Excitation Objective, 20x/0.60 NA, 5.5 mm WD - Multi-immersion objective, 10mm WD, NA 0.7 @ RI 1.45, EFL 8.4mm @ RI 1.45. (R.I. range 1.33 to 1.56) At RI = 1.33, NA = 0.64; XY resolution = 470 nm, Z resolution = 3.2 um - dual Hamamatsu ORCA-Fusion (back thinned) sCMOS cameras with CameraLink frame grabber cards (i.e. 2 fluorescence channels simultaneously) - ASI typically uses micro-manager.org to control all the components on th light sheet sub-system (which in turn would be triggered by Hudson Robotics). - GM anticipates we will have GPU deconvolution at some point in workflow (maybe not 12/2022 "first light". - PC - nice workstation (including 10 Gbe Ethernet). * Liconic STR 44 incubator to hold up to 44 plates (i.e. 37 C, 5% CO2) for hiPSCell organoids.
|
|
Feature(s): tile scanning & image stitching and/or stage relocation Note: four of our microscopes can "do" tile scanning and image stitching AND/OR automate stage relocation for "cyclic" imaging (ex 12plex RNAscope 3 cycles of 4 probes)(save position list, reload when reutrn to slide - assumes you position the slide or imagign dish consistently). This can enable more efficient acquisition at optimum spatial resolution (i.e.high NA objective lens, wavelength limited pixel and Z-step size) (if the analysis computer can handle our 'data deluge') without you being present the whole time (a major benefit of automated microscopy): * ACCM Leica SP8 confocal microscope (standard tile scan or NAVIGATOR 'app' wizard) * Olympus FV3000RS confocal * FISHscope (Olympus IX83) * Keyence BZ-X710 Our -- now obsolete and dismantled -- Olympus FV1000MPE multiphoton microscope had tile scan -- a user 'did' a 50 slide project of kidney tissue fibrosis in 2018 -- but currently (1/2021) very nfrequently used microscope (we'd be happy to see more users on it). ==> Please note Olympus is now known as Evident Scientific. 1/2021: we have also put on VueScan slide scanner for histology / brightfield IHC slides onto a PC in the image core and an iLab scheduler here. This produces images equivalent to a 4x or 5x objective lens 'nominal resolution' (though sometimes the slide scanner focus is not quite as good). Nice for acquiring large area quickley, and then making measurements of areas in Fiji ImageJ, MetaMorph, Photoshop, etc. If users find it useful, great; if not, we may remove it as a service and just have it at GM's PC. Scanner is a repurposed 35 mm film scanner. You are welcome to scan in your 35 mm film 'cassettes' or film strips, when VueScan is not being used for research. |
|
Manual stitching tips (can also be useful if the acquisition machine stitching does not work perfectly or is not "time effective" to do at the acquisition PC) ... * when possible, move in one axis only; watch 'marker' as you move and stop before the marker goes out of view on the other side. * Typically you should acquire with 10% or more overlap of tiles. Sometimes 20% -- i.e. test 10% vs 15% vs 20%. You might put small pieces of sticky note(s) on the monitor as a visual guide for how close to the edge(s) 10% 915%, 20%) is. * Photobleaching is bad: You may want to turn down the epi-illumnation intensity (the "big dial" on the Zeiss inverted microscope lamp ... be sure to reset the dial to 100% for the next user) and longer exposure time. Photobleaching is "non-linear": higher intensity illumination causes disproprotionally more photobleaching. * Photobleachign is under your control with sample preparation: use the right mounting medium [consider testing Prolong Glass, buy without DAPI; if need DAPI, put it in with primary and/or secodnary antibodies, and wash extensively to get black background outside cells/tissue] and right fluorophores; Tip: Tyramide signal amplification (TSA) can make fluorescence signal 100x brighter than standard immunofluorescence, enabling shorter imaging time (effectively no photobleachng). Also possible to multiplex TSA. * Sparse specimen stitching: most stitching software relies on "features" to stitch, so if very sparse, may nto work. * Zero overlap stitching (automated microscopes): the field of view can be known precisely (from image settings) so you COULD acquire ADJACENT fields of view and use MetaMorph's "Montage Stack" to stitch (limit: 30,000x30,000 pixels, which is pretty big). We have Metamorph on severl PCs. *** Liu JT 2021 Harnessing non-destructive 3D pathology . Nature Biomedical Engineering (Feb 15, 2021 epub). https://doi.org/10.1038/s41551-020-00681-x Image stitching. A key step in the acquisition pipeline for 3D microscopy data is the efficient assembly of large numbers of 2D image tiles into seamless volumetric datasets. A number of commercial software packages (such as Imaris Stitcher and Volocity) as well as popular open source tools (such as TeraStitcher ref 142,143 and BigStitcher ref 144) have been developed to address this challenge. Some software tools (in particular, BigStitcher) are designed to correct for deformation and registration artefacts through affine transformations, including chromatic shifts between wavelength channels. ==> GM notes: * QuPath https://doi.org/10.1038/s41551-020-00681-x (Bankhead 2017 Sci Reports) does not appear to have image tile stitching, may still be of use for image analysis. * Adobe Photoshop (CS1 and later) Automate/Batch has image stitcher. * Microsoft Research - Image Composite Editor (ICE 2.0) free image stitcher ** Feb 22, 2021: "Work in progress" - we have set up a histology/immunohistochemistry brightfield slide scanner, we call "VueScan". Image quality is about the same as a microscope 5x objective lens, but "VueScan" acquires a much bigger field of view in under 2 minutes. 5x lens ~2x2 mm, VueScan ~36x24 mm. You can then use the Vuescanned images to guide your imaging on our other microscopes (i.e. upload to your Microsoft OneDrive, and either print out images or organize your project into PowerPoint etc, download to the microscope PC or bring your own laptop, and reference the "overview" data). VueScan is in iLab - please contact George for training. The "VueScan" is named for the image acquisition software (Hamrick VueScan Pro, www.hamrick.com), the scanner is a Plustek OpticFilm 8200ai 35mm film scanner with one of the film strip holders modifed by George to hold a microscope slide. I also note that if you have a flatbed scanner in your lab, ideally with transparency lid, you could acquire in your own lab (VueScan would be good software to use). |
|
Scientific Image Ethics - a few links ==> does an instrument/services page seem like a weird place to put a section on image ethics? No! If you want to use our core, please plan to, and do, behave ethically. JHU has an online data repository - for journals that do not enable you to (cost effectively) post all your data as "supplementary" information 9in original data format, with MetaData, and your complete methods, including any image processing and analysis 'scripts/macros/journals'), you could post it at CRAEDL, and include the DOI with your paper. Hopefully they do - or will -- enable preprint data to be posted. preprints both give you academic credit (that is, you have a date you can claim a discovery) and nable getting feedback on your work. See also ELife's 2020 editorial on "moving to preprint reviewing" model, https://elifesciences.org/for-the-press/a4dc2f54/elife-shifting-to-exclusively-reviewing-preprints https://craedl.org/docs 20241206F: Craedl is no more. See JHU Research I.T. SafeStor https://researchit.jhu.edu/safe-stor/ *** HHS (NIH's parent organization) Guidelines for Best Practices in Image Processing 2008 https://ori.hhs.gov/education/products/RIandImages/guidelines/list.html (web page has live links to Photshop videos for each item)
*** which was cited by: Brocher 202102 BioVoxxel preprint - Seeing the Big Picture - Scientific Image Integrity under Inspection - Project Image Integrity Assessment * https://www.researchgate.net/publication/349521202 *** I am a big fan of Jerry Sedgwick's (2008) section on microscopy image ethics in: https://www.amazon.com/Scientific-Imaging-Photoshop-Measurement-2008-06-02/dp/B01JXNG5IK Scientific Imaging with Photoshop: Methods, Measurement, and Output by Jerry Sedgewick Notes: * don't pay a crazy price for the book (post covid-19 andemic you could borrow mine). The JHU library might have a print you could check out. * disclosure: I wrote a favorable review/comment of the book on Jerry's request.
|
PIs: to set up a new user and cost center, see our iLab instructions page.
Monday June 15, 2020 we re-opened as part of JHU-Research's covid-19 phase 1 re-opening.
All users must read - and follow - our Covid-19 Phase 1 re-opening policies
http://confocal.jhu.edu/covid-19-policies
Quick summary:
1. Obey room occupancy limit (posted at door). 12/2020: 1 person per 200 square feet, so small rooms occupancy = 1.
2. wear your face mask (cover nose and mouth), and new lab gloves (bring your own supply) when touching microscope, mouse, keyboard, etc. If you handle a specimen for biosafety reasons, discard those gloves in biosafety trash and put on new lab gloves for microscope etc.We provide 'saran wrap' at each station - you are welcome to discard old wrap and apply new wrap at the start (and if needed during) your session. typically keyboard, you can also cover mouse, eyepieces.
***
** Please note that image core management sometimes has to cancel/postpone user sessions due to required service visits or other reasons. When this happens we will try to make the user's next imaging session be "no charge" (even if longer than the cancelled session).
Please Reserve Your Instrument!!! Ideally 24 or more hours in advance; if not in advance, then right after you fill in the paper sign-in sheet. Reservatons policty is online at
http://confocal.jhu.edu/reservations-policy
Please read - and follow - our Biosafety Policy
http://confocal.jhu.edu/biosafety-policy
|
Quick biosafety in image core tips: * "safety is job #1". If you are not using our equipment in a safe manner, then do not use our equipment. * do not touch our microscopes,keyboards, computer mice, door knobs, light switches, etc, with lab gloves. * Yes, handle your SBS plate, imaging dish, other biohazard specimen, with a new lab glove(s) ... after adjusting -- without glove -- the microscope condenser arm, etc, to recieve/remove your specimen. * Yes, biohazard spills have happened here at JHU/JHMI - see July 2018 tuberculosis spill national news. * Transport your specimen(s) in a spill resistant tray(s) and/or box(es), i.e. on a lab cart. Do not touch the passenger elevator door buttons, or stairwell handles, with lab gloves. * our major users include 6 different models of infectious diarrhea, and potentially clinical specimens from G.I., with "C. diff" and/or novel infrectious agents, and Prof. Cindy Sears lab, who have shown that essentially all human colorectal cancers involve contributions from infectious agents (800,000 new cases per year woldwide). It is critical that all users conduct experiments in a manner that is safe for both the user, other users, core staff, custodial staff, and visitors. * Dr. McNamara is the image core manager and the biosafety officer for the microscope rooms. If you are3 unwilling to follow our biosafety rules, either you and your P.I. should explain to Dr. McNamara and Image Core Director Prof. Bin Wu, how to improve our biosafety policies, or you should not do experiments here. |
***
Data transfer:
Plan A (best): your MyJH -> your Microsoft OneDrive "JHOneDrive": Nearly all our PC's can see "The Web" and so can see MyJH. Now (mid-2018) that every JHU staff member gets 5 Terabytes of Microsoft Onedrive "cloud" storage space, you can transfer from the acquisition PC or our server to your OneDrive account. Be sure to logout ("sign off") of your OneDrive session here when you are done -- you need to do this manually. You also need to log out of your MyJH session.
Plan B: Many JHU SOM computers can see our file server. This is the simplest way to transfer files for those with access. Do not give out the name or I.P. address of our server. We prefer you use your JH OneDrive (plan A above).
Plan C (risky): You / your lab could 'expose' some "share folder" on your PC or server and transfer from our PC/server to your share. You should only do this with explicit permission of your P.I. and network administrator, since setting up your 'share' incorrectly could expose your PC/server to attack.
NOT Permitted: USB drives are NOT permitted on our computers. We will dispose of them in the Biosafety trash.
***
Legacy equipment (retired) include:
Andor spinning disk confocal microscope. ... this has been moved to a research lab. Please contact George (gmcnama2@jhmi.edu) if you want to get in contact with the P.I. who has the Andor.
http://confocal.jhu.edu/current-equipment/andor-spinning-disk
Zeiss AxioImager.M2 automated upright microscope, with PTI DeltaRAM high speed random access monochromator, RatioMaster software, MetaMorph license #4646 (repurposed for analysis), Photometrics Coolsnap HQ CCD camera (nice in early 2000s, now outclassed by sCMOS cameras such as the ORCA-FLASH4.0LT on our FISHscope), standard metal halide lamp (interchangable with DeltaRAM). Was used for Fura-2 ratio imaging Ca++ ions - a method mostly obsolete with GCaMP6 and similar fluorescent protein biosensors and image on our Olympus FV3000RS confocal microscope, with resonant scanner mode available 512x512 pixels (1 fps at 15 line average), also "kymograph" line scan acquisition. This microscope does not have any environmental control unit.
http://confocal.jhu.edu/current-equipment/zeiss-upright-scope-w-pti-ratiomaster
Olympus FV1000MPE - multiphoton microscope retired (and as of early 2025 dismantled and removede) due to failure of a critical component for controlling the intensity of the laser. The cost of the component and high probability of other part(s) failing, have led us to de-commission this microscope. The laser still worked (when I turned it off Spring 2022) and some objective lenses still useful (we might even move some to FISHscope). We are open to discussions on re-purposing the laser and/or other components.
==>Please note Olympus is now known as Evident Scientific.
***
As of July 2022 Dr. McNamara works with Prof. Jeff Mumm, Wilmer Eye Institute (WEI). Most of the user's are in WEI, and there are a lot of other confocal microscopes on campus (including WEI's core facility), so probably not needed by other researchers. Very brief summary:
* Olympus upright microscope stand, FV1000 FluoView software (old generation - different from the FV31 "FluoView" software on our FV3000RS). Windows XP PC so not on campus network.
* laser lines 405, 440, 488, 515, 559, 635 nm (6 laser lines, added to system over several years as need and money arise).
* five PMTs: CHS1, CHS2, CHS3, HSD1, HSD2 (CH = channel, "S" = standard PMT, HSD = High sensitivity PMT) + "TD" transmitted light detector
.
***
Vendors contact information for our instruments is online at http://confocal.jhu.edu/vendors-contact-information
-
. FISHscope
FISHscope - use iLab (currently limiting to select users)
Location: Ross Imaging Core (Ross Bldg 9th floor)
-
. Leica SP8 confocal microscope
Leica SP8 Confocal Microscope (owned by ACCM, open access)
Location: Ross 9th Floor
-
-
. Olympus FV3000RS Confocal Microscope_part2
. Olympus FV3000RS Confocal Microscope_part2
Location: fv3000_part2
-
ACCM_IncuCyte_S3
Live cell imaging system inside 37 C incubator - short to long term (many days possible) live cell imaging flasks and SBS plates (can hold up to 6)
Location: Ross3
-
-
-
-
MetaMorph Key 2 (#4646) (AxioImager Upright license)
July-August 2017: we are considering transitioning to keep this on one microscope/PC
Location: S972
-
- 1 2 >