Keyence BZ-X700 (open access)

Keyence BZ-X700
Location: Ross 9th floor
Keyence BZ-X700 with Tokai Hit Incubator
20240815Thur: this week a user showed GM the "red filter cube" images are dimmer than in the past. The other cube images look ok. GM's conclusion: bizarre issue (unless a user wrecked the cube). Mybe you should move to FISHscope (gget training).
20210107F: we anticipate more use of Keyence for fluorescence slide scanning - now that we have a good work flow for "shading correction". Key to speed: is scan one fluorescent channel, then change channels, scan again. Stitch in the Image Viewer software (with shading correction). Usually use "standard" acquisition (2x2 binning, 2x gain).
Keyence can also scan H&E, "special stains" and other brightfield slides.
On JHU SOM campus, you can contact Comparative Pathobiology for getting tissue sections cut and stained, such as H&E, etc, https://mcp.bs.jhmi.edu/histology-price-list/
|
20250401U - RGB with Cubes in place McNamara 20250401U – Brightfield – maybe even RGB or RGBNir – with all cubes in place RGB: You COULD set the blue, green and red cubes to brightfield, acquire all three. ==> if tile scan, please do sequentially to minimize wear and the on the filter cube turret – then combine in Photoshop or Fiji ImageJ etc. (... "sequential" is one cube at a time, so 3 or 4 scans, which is also a lot faster than "simultaneous" multi-channel. RGBNir: For that matter, you could also acquire brightfield through the Cy5 cube – H&E (or H-only, E-only slides) are “practically transparent” in the near infrared, so gives you an unstained slide image. George p.s. this is probably worth publishing. Best wishes on that! (preprint then peer review). From: Willem Buys <wbuys1@jh.edu> Hi George, Usually, when imaging Brightfield on this microscope, I set it to the Cy5 channel and take the filter cube out, as you have showed me. ==> Is there a possibility to image Brightfield with the filter cube still in? ==> And would that work in a channel other than Cy5, which is already in use in my experiment? Otherwise I'll have to find the area of interest in the Brightfield and then put the cube back in before imaging with Fluorescence. Thanks for your advice and best wishes, Willem |
|
McNamara 20220420W - Keyence Stitching Tips * Make sure your slide is as flat as possible … slide holder seated correctly on the stage, make sure slide is on correctly (not raised on a “shelf”), no slide label or label as thin as possible.
* Big stitched images may not be viewable with common viewers. MetaMorph limit 30,000x30,000 pixels. The simple (free, you get what you pay for) Microsoft Photo Viewer is mediocre. Fiji ImageJ has limits too. GM often uses IrfanView (www.irfanview.com – Windows 64 bit) for TIFF, JPEG, etc (it can let you reduce size to save 10%, then save to a new filename). * We do not usually have a Nikon 40x lens on the Keyence. We may be able to borrow one from a Ross 9 lab. Plan in advance (if you would like to buy us a nice Nikon lens, lets talk: “lens for time”). |
20191104M (November 4, 2019): we are now on JHU iLab for scheduling. All users need to reserve in iLab AND sign in on paper sheet (and sign out when done). We use iLab reservation for billing (and spot check with sign in sheet. We remind users that using the machine without sign in is trespassing - which JHU could fire you for.
Ross Imaging Center
If a user needs their PI or administrator to set up account numbers (IO#'s in JHU accounting jargon), see
https://help.ilab.agilent.com/36900-managing-your-group/279959-membership-requests-fund-numbers
** Please note that image core management sometimes has to cancel/postpone user sessions due to required service visits or other reasons. When this happens we will try to make the user's next imaging session be "no charge" (even if longer than the cancelled session).
***
Nikon objective lenses CCD camera 1920x1440 pixels
4x/0.13NA PhL 3623x2717 um, 1.88 um pixel size
10x/0.30NA Ph1 DL 1449x1087 um, 0.75 um pixel size
20x/0.45NA Ph1 ADM 725x554 um, 0.38 um pixel size
Keyence camera 1920x1440 pixels at "high resolution" (1 pixel) setting ... also has (if I recall correctly) 2x, 3x, 4x, 8x (12x?) binning, resulting in "superpixels" -- lower resolution, higher sensitivity because a superpixel is readout one time, not (if 12x12) 144 times. The Keyence uses CCD sensor, so can do this. Most 2025 era cameras are now sCMOS, which can only bin in software post acquisition.
Special projects only: 100x/0.145NA oil immersion (not kept at microscope). 145x109 um, 0.076 um pixel size.
| Fluorescence | Excitation | Dichroic | Emission |
| DAPI or Hoechst | 360/40 | 400 | 460/50 |
| Green ... usually Alexa Fluor 488 | 470/40 | 495 | 525/50 |
|
TxRed, Alexa Fluor 594, Alexa Fluor 610-X |
560/40 | 585 | 630/75 |
| Cy5, Alexa Fluor 647, ATTO647N | 620/60 | 660 | 700/75 |
* Metal halide lamp.
* Brightfield is normally done through the Cy5 filter cube (software configuration option) (avoids users having to add/remove cube)..
* TxRed is the name of the filter set in the filter cube. It is named for Texas Red. Many users use Alexa Fluor 594 instead.
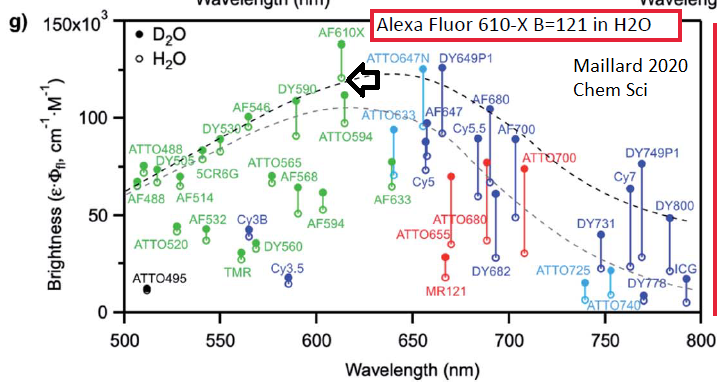
==> In late 2021 GM found a paper (Maillard 2020 Chem Sci - see multiplex web page) that reports Alexa Fluor 610-X is more than 2x brighter than Txas Red, AF594 or Cy3.5.
Maillard article https://pubs.rsc.org/en/content/articlelanding/2021/SC/D0SC05431C key figure (open circles are for water, H2O ... while you can buy deulterated water, D2O, may not be practical).
* Best practice stitching: ONE CHANNEL at a time. This avoids changing filter sets during the tile scan. For example, 75 min for 2 fluorescnce channels (4x objective lens, ~20x50 mm area, i.e. 2/3rds of a slide) vs 7 minutes for one channel, change to next filter set (orange background button of filter cubes at top-of interface), then another 7 minutes.
* Stitching in the Keyence software - sometimes results in annoying "stripes". these "stripes" can be removed using shading correction in the Keyence "Image Viewer Wide" software ("analyze" button). George can show users. It is important to be consistent with camera settings -- ideally use "Standard" (2x gain, bin 2x2) -- to avoid needing to make (for yourself) shading reference images for every possible channel * objective lens * camera gain * binning (4 * 3 * 5 * 6).
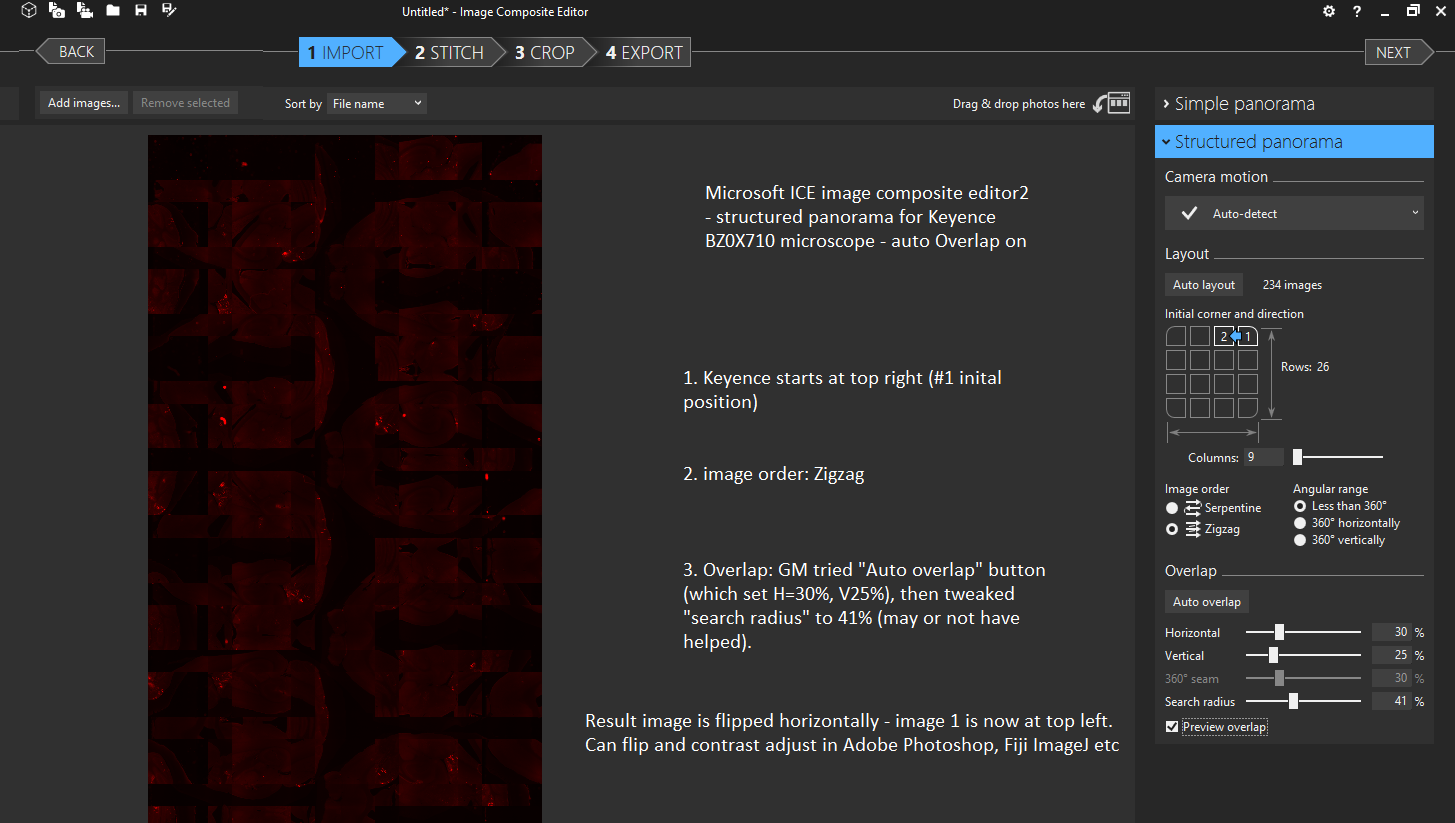
GM found settings for Microsoft Research ICE 2 (Image Composite Editor) that worked better:
***
Specimens (keep your specimen clean! both in tissue culture incubator, hood, lab bench, Keyence work area):
* microscope slide (coverglass down toward objective lens standard).
* SBS plates ... standard plastic tissue culture plates work, "imaging quality" plastic fine, better image quality piossible with COP ~170 um thick bottom, glass bottom (ex. mattek) high quality (but pricey).
* 35 mm plastic tissue culture dish ok, coverglass bottom (170 um) may be better.
37 C, 5% CO2, humidity available. Timelapse interval depends on number of fields of view and acquisition settings.
For SBS multiwell timelapse imaging projects we suggest starting with 24-well SBS plate, fill all outer wells with dH2O (or sterile PBS), use the inner 8 wells (4x2) for experiment.
Keyence is motorized XY and Z. Typical uses are:
A. select field of views of slide/well(s),
B. tile scan, stitch later.
note: always save to the Keyence PC (not any network drive), transfer to your JHU Microsoft OneDrive at end of your session (and log out of OneDrive and myJHU).
***''
==> multiple fields of view --> stitch ... alternative: our Zeiss Axio Observer.A1 Inverted Microscope now has "manual MIA" stitching in Olympus CellSens. I note that both Adobe Photoshop CS__ and the free Microsoft ICE (Image Composite Editor) can stitch. Fiji ImageJ also has various stitchers.
Keyence Access Policy (20180914 update) -- As of 20190625 will be strictly enforced.
- The Keyence microscope is owned by two labs and managed by the image core. The two labs -- Prof. Mark Donowitz (G.I.) and Prof. Peter Andersen (Cardiology) have priority access. As of now, this means members of these two labs can make reservations 2 week in advance of their session start. All other users should reserve less than two weeks in advance. We note that occasionally experiments are done for "many days" (ex. 8 days) - we ask such users to schedule gaps of "a couple of days" between replicates. For example: 8 day run - 2 work day gap - 8 day run - 2 work day gap - 8 day run.
- Multi-hour experiments: when possible, should be conducted overnight (by fully trrained users), not take up significant workday hours (ex. 4pm Tue -> 9am Wed)..
- Multi-day experiments: when possible, should be conducted over weekend (ex. 4pm Fri -> 9am Mon).
20181018 update: Ryan Parent is primary Keyence contact (Ari Klein moved to Florida) and Takanori Tsuchiya is primary Tokai Hit contact (Sato-San returned to Japan).
|
Ryan Parent RParent@keyence.com Sales Engineer Life Science Microscope Team Keyence Corporation of America 700 American Ave Suite 100 King of Prussia, PA 19406 |
|
Takanori Tsuchiya |
20170808: Sato-San from Tokai Hit installed a new incubator lid (a user broke a cable of the previous lid). All users should use the Tokai Hit incubator inserts when using a microscope slide or 35 mm dish (holds 2 dishes, if using only one, put an empty dish on other opening to keep the humid air in the incubator volume, not into the Keyence body) or 50 or 60 mm dish holder (please contact George if you want to use the 100 mm dish holder that came with the Keyence BZ-X700 microscope).
* SBS plates
* microscope slide
* one or two 35 mm plastic dishes or imaging dishes the high NA objectives require use of imaging dish). *** if using one dish with humidified air, cover the other opening with a dish.
* 50 or 60 mm plastic or imaging dish (this holder can also accept a standard 75x25 mm microscope slide).
My special thanks to Sato-San of Tokai Hit for excellent support and training.
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Glass bottom dishes and SBS plates can be purchased from Mattek, https://www.mattek.com/store-category/cultureware/glass-bottom-dishes/
Most users should order "1.5" thickness coverglasses (specification refers to manufactururing in 160 to 190 um range); the "0.170" millimeter thickness = 170 um are higher precision. Several other vendors also offer imaging dishes and with a dentist circular saw and silicone cement (a.k.a. aquarium glue sealer), I used to make my own (and still have some 40x50 mm coverglass in 100 mm diameter petri dishes).
Nice information on coverglasses:
http://www.hausserscientificglass.com/products/precision-glass.html
Size Thickness (inches) Thickness (mm)
#00 0.0020-0.0033 0.05-.008
#0 0.0033-0.0051 0.08-0.13
#1 0.0051-0.0063 0.13-0.16
#1 1/2 0.0063-0.0075 0.16-0.19
#2 0.0075-0.0098 0.19-0.25
#3 0.0098-0.0138 0.25-0.35
#4 0.0170-0.0250 0.43-0.64
I note that Hausser Scientific [without glass in name] offers 0.4 mm and 0.5 mm thick coverglasses; I included 1.0 mm, which is standard microscope slide thickness:
http://www.hausserscientific.com/products/cover_glasses_slips.html
Catalog # Description
1461 20mm x 26mm x 0.4mm
5000 20mm x 26mm x 0.5mm
5010 25mm x 28mm x 0.5mm
5020 25mm x 33mm x 0.5mm
5030 27mm x 37mm x 0.5mm
5040 Sedgewick Rafter cover glass 25mm x 60mm x 0.5mm
5051 Petroff-Hausser 20mm x 26mm x 0.2mm
5060 Whipple eyepiece micrometer 21mm dia.
5075 Graduated Howard Mold Cover Glass, Unlaminated
5075L Graduated Howard Mold Cover Glass, Laminated
5080 Howard thin 28mm x 33mm x 0.5mm
5090 Howard thick 28mm x 33mm x 1.0mm
5400 Nageotte 30mm x 33mm x 0.5mm
5411 Nageotte 30mm x 33mm x .17mm (1 oz box) disposable
Reserve Equipment

