. FISHscope

FISHscope - use iLab (currently limiting to select users)
Location: Ross Imaging Core (Ross Bldg 9th floor)
FISHscope
Main application: single molecule RNA fluorescence in situ hybridization (smFISH).
FISHscope Quick Tips web page (startup, finish session) is at http://confocal.jhu.edu/mctips/fishscope-quick-tips
Our thanks to Prof. Mark Donowitz for supporting our NIH NIDDK P30 grant (P30 DK089502) supplement application and matching funds, and to NIH -- U.S. taxpayers !!! -- for providing the supplement $. Please cite the grant number in your acknowledgements.
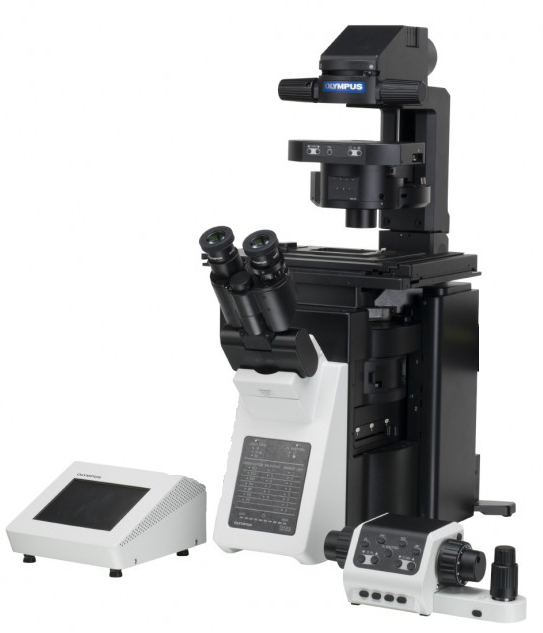
Olympus IX83 inverted microscope running Olympus cellSens software.
==> Please note Olympus is now known as Evident Scientific.
If you want to provide more details, choose any or all of:
Olympus IX83 inverted microscope running Olympus cellSens software, Lumencor SPECTRA-III 360 eight channel light engine. Semrock LED-DA/FI/TR/Cy5/Cy7-A penta cube, Sutter 10-3 emission filter wheel with Semrock 32 mm diameter filters, Hamamatsu ORCA-FLASH4.0LT sCMOS camera. Olympus UPLSAPO 60x 1.35NA oil, UPLSAPO 20x 0.75NA, SP 10x/0.40NA, UPLSAPO 4x 0.16NA.
Our thanks to John Gibas, [now ex] Olympus Corp., Peter Brunt (AVR Optics - Semrock's distributor), Erich Zeiss and Iain Johnson (Lumencor) for working with us and our other vendors, for getting this instrument specified and delivered.
Typical Experiment uses these five fluorophores and our "Penta" filter cube ("4plex plus DAPI")
* DAPI
* Alexa Fluor 488
* Alexa Fluor 568 (Penta cube is better match to AF568 than to others - see also below).
* Alexa Fluor 647
* Alexa Fluor 750
optional plex: ** Alexa Fluor 700 filter set arrived 12/2022 (users will need help from GM to update Process Manager to use this cube).
** we can still acquire using our "R10/T90" beamsplitter, this AF700 cube will be better (faster, lower background). to enable "optimized 5plex + DAPI". (ex. AF750, AF700, AF647, AF555, AF488 + DAPI).
optional plex Alexa Fluor 610 filter set ... the mCherry filter set was moved from the FV3000RS confocal microscope to FISHscope and renamed for AF610, Alexa Fkuor 610 (see Maillard 2021 Chem Sci, Brghtnesas in H2O 121, in D2O 132, highest of 42 dyes tested in solution), Texas Red (quantum yield 0.93), Alexa Fluor 594, nCherry, other Red fluorescent protein (mScarlet3 etc),
upshot: FISHscope can enable you to do" 7plex + DAPI: example, AF750, AF700, AF647, AF610, AF555, AF488 + DAPI. Potentially additional plex with R10/T90 beamsplitter (cellSens Process Manager can have maximum 20 settings). Fun fluorescence fact: if you have spatially separate "features" could do "combinatorial labeling", where antibodies or FISH probes are labeled with 1 or 2 or 3 etc fluorophores. Spectral Karyotyping (SKY) uses 5 fluorescent dyes in combinations of 1, 2, 3 or 4, ex. A,B,C,D,E ... AB, AC ... ABC, ABD ... ABCD, ABCE, BCDE, generate 24 probe sets, label each of the human chromosomes (22 autosomes, X, Y sex chromosomes).
For those of you using Li-Cor ODYSSEY western blot scanner or similar NIR fluorescence instruments, FISHscope MAY work for IRDye680 or IRDye700 (use one or the other) and/or IRDye800. These are sometimes used on direct labeled "clinical" antibodies in mouse tumor xenograft in vivo fluorescence imaging. Please be sure to bring positive controls (in vitro labeled, extensively washed cells, and appropriate tissue section), negative controls (no IRDye800 antibody; potentially "isotype control"; ideally antigen knockout mouse control tissue and/or pre-incubate with unlabeled antibody, then again with unlabeled + IRDye800 labeled).
* Cy5 and Alexa Fluor 647 channel ... negative control (might have weak IRDye680 or IRDye700 crosstalk)
* Cy5,5 and Alexa Fluor 700 channel ... IRDye680 or IRDye700
* Cy7 and Alexa Fluor 750 channel ... IRDye800
|
* Semrock LED-DA/FY/TR/Cy5/Cy7 ... DAPI, Fluorescein (Alexa Fluor 488), TRITC (not Texass Red, not Cy3, not Alexa Fluor 610), Cy5 (Alexa Fluor 647), Cy7 (Alexa Fluor 750 ... often superior to IRDye800) * Semrock AF700 ... new (12/2022) Alexa Fluor 700 filter cube * LED-mCherry ... Alexa Fluor 610 (and mCherry, Texas Red, Alexa Fluor 594, etc) ... move from FV3000RS (since Redundant with TRITC cube "orange" fluorescence for looking by eye) * R10T90 (R10/T90) and R03/T97) reflection/transmission beamsplitters in cubes ... flexible choice of Lumencor SPECTRA III-360 lamp channels and emission filters in Sutter wheel. |
Request: we have a limited budget so no money to do this: please fund our purchase of a dedicated Alexa Fluor 610 filter set (details "to be determined"). to enable "optimized 6plex plus DAPI". ... GM looking at Semrock Searchlight graphing web site https://searchlight.semrock.com/ THINKS we could image EACH of Alexa Fluor 546 (or AF555) AND Alexa Fluor 568 AND Alexa Fluor 610 separately (at least in context of single molecule RNA FISH "dots") if we had THREE optimized filter cubes. However, at approximately 3 * $1800 = ~$5400 (maybe as much as $6000) this is a non-trivial cost (The IX83 microscope stand filter cube turret has 8 positions, now (or soon): 5 cubes = Penta, R10/T90, R03/T97, Alexa Fluor 700 (AF700 cube added 12/2022), CaliCube, so could take another 3 without bumping any (we rarely use R03/T97 with R10/T90 being similar).
This microscope specifications were developed by Prof. Bin Wu (Image Core director) and George McNamara, PhD (Image Core Manager), for single molecule RNA FISH probe sets developed by Prof. Bin Wu's lab (more on this below), and is also capable of multiplex immunofluorescence (including Brilliant Ultraviolet BUV395, Brillian Violet BV421, and their tandems), combined smFISH and immunofluorescence, and commercial smFISH probe sets from vendors such as Biosearch Technologies ("STELLARIS FISH)") and ACDBio/Biotechne ("RNAscope") ... also rolling circle amplification (RCA), hybridization chain reaction (HCR), etc. We recommend communicating with George McNamara about any unusual reagents (such as non-standard fluorophores). George also tries to keep up with "fluorescence literature", for example, Alexa Fluor 610-X is reported to be ~2.4x Brightness (E.c. * QY / 1000 = 121) vs Alexa Fluor 594 (B = 54) (http://confocal.jhu.edu/mctips/af610-x).
|
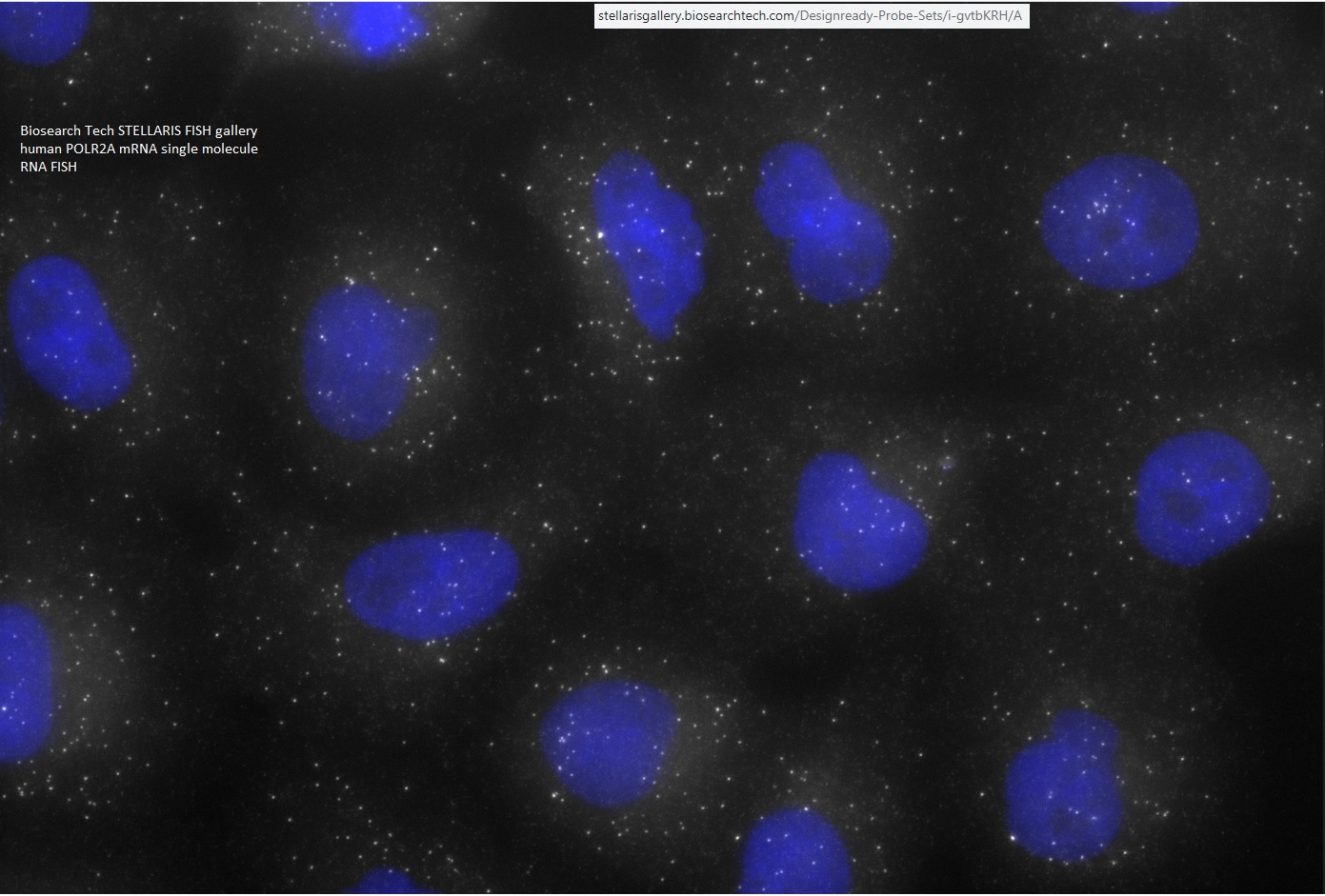
FISHscope is aimed at producing high quality fluorescence images: * multiplex across spectral range (DAPI, BUV395, BV421 ... Alexa Fluor 750, Cy7, IRDye800 etc) * "dots" (puncta, sub-resolution spots), such as fluorophore-oligonucleotide pool based single molecule RNA smFISH (Raj 2008 Nat Meth - pubmed 18806792), RNAscope (ACDbio/bioTechne, also available from Panomics/Affymetrix/ThermoFisher), Hybridization Chain Reachion "HCR" (https://www.molecularinstruments.com -- also various academic labs have made their own variants) ... also "TattleTALEs" concept of GM ( https://works.bepress.com/gmcnamara/75 -- more focused on live cell imaging ). Why widefield usually outperforms confocal microscopy on smFISH [see after the 2nd paragraph for 3 line summary, numbers in the first 2 pararaphs]: FISHscope (widefield) uses an sCMOS camera with 4 million pixels (2048x2048 pixels). Often for smFISH probe set(s) we use a 4 second exposure time (sometimes much shorter), and do Z-series with 63x/1.35NA objective lens, 0.3 um step size, ~10 um, so ~30 planes, so 120 seconds per channel (and then 3D deconvolution, which takes more time, but usually worth it). the cameras has a peak quantum efficiency of ~80% (QE curve not flat, but not critical for this discussion). Illumination is widefield (ours has 6 LEDs, 2 laser lines). We think we are "soemwhat below" fluorophore saturation for each excitation channel (we normally operate the Lumencor SPECTRA-III 360 lamp at 25% power to avod saturation). We deliberately chose 60x/1.35 NA (budget limited number and quality of objective lenses -- can put same Olympus lens on FISHscope IX83 stand and FV3000RS IX83 stand) and 108 nm XY pixel size that is "somewhat larger" than theoretical optimal size for pont spread function. Confocal -- I use Olympus FV3000RS as example, our Leica SP8 is similar -- has user selectable image formats, including 2048x2048 pixels (match FISHscope camera) and variable zoom, so we can match image dimensions AND pixel size (if we want). We find scanning at maximum speed in "galvo" mode to give best data, due to minimal photobleaching compared to slower scanning settings (my thanks to Jonathan Boyd, PhD, now ex-Leica, for demonstrating the impact of scan speeds on photobleaching). I always use fastest scan speed, 2 microsecond pixel dwell time ("2 usec PDT") and some averaging. For here, to simolify math, I use 10 line averaging (max on FV3000RS galvo mode is 16 averaging ... could use deliberately small Z-step size to effectively average more times). The maximum quantum efficeincy of FV3000RS GaAsP PMT detectors is ~40% (probably less, but 40% simplifies comparison). We have seven laser lines, each of which can deliver sufficient power -- if we want -- to photobleach any fluorophore (also in play is what zoom is used: 10x zoom concentrates 100x power into the field of view). We deliberately avoid using enough laser energy to photobleach! However, we probably are still "close to saturation" of whatever fluorophore(s) absorb at or near the laser line. ... Our Leica SP8 confocal microscope has better detectors (2nd gen HyD) that operate in photon counting mode, but similar calculation as for FV3000RS. widefield FISHscope: 4 seconds per pixel, 80% quantum efficiency. Confocal (FV3000RS); 20 microseconds per pixel [0.020 seconds], 40% quantum efficiency. Laser focused to resolution limited spot, probably "close to" fluorophore(s) saturation (laser settings usually tuned to avoid photobleaching). So: 4 sec vs 0.020 sec = 200x longer exposure, 80% vs 40% (optimistic) quantum efficiency = 2x advantage, so FISHscope is collecting 400x more photons than FV3000RS if same "excitation photon flux" (or percent fluorophores excited and result in fluorescence photon emission in point spread function spot) and assuming no photobleaching. Tips to optimize either method: * start with thin tissue culture cells to gain experience ... good results --> further optimizations of procedures and reagents, less likley to confuse researchers into 'wishful thinking" (in replication studies, ~50% of published experiments were not replicated [50% gave more or less similar results, though usually with lower p-value statistical significance], implying original researchers has some or a lot of wishful thinking ... and likely went on to further studies that might not be replicated if tested]. * use DAPI or BV421 antibody for "blue" channel -- high autofluorescence background makes smFISH in this channel difficult. * use "green" channel" to image GFP or Alexa Fluor 488 antibody, because if try to go FISHing in this channel may get "puncta autofluorescence" which could be confused with FISH dots. * still leaves at least 4 "channels" for FISH (on FISHscope, maybe 5 more) -- in Biosearch Tech fluorophore terms, Quasar 570 [Cy3 like], CAL Fluor 610 [Cy3.5, Alexa Fluor 610-X like), Quasar 670 (Cy5, Alexa Fluor 647 like), Quasar 705 (Cy5.5 like) ...GM does not have personal experience with this fluorophore. Alexa Fluor 750 (Cy7 like) Biosearch Quasar's: https://www.biosearchtech.com/support/education/fluorophores-and-quenchers/quasar-dyes * Uusually confocal microscopes are operated with the confocal pinhole set to 1.0 Airy Units ("1.0 Airy Disk" in FV3000RS). Larger pinhole -- I suggest you could try 2.0, 3.0, 4.0 Airy Units (may need to reduce laser power to avoid detector saturation), followed by spatial deconvolution, may result in more useful data. Larger pinhole setting(s) collect more light. I note that Olympus states that on the FV3000RS that 500 Volts is optimal "HV" setting for the GaAsP detectors (max is 900 V, which is likely "single photon sensitive" but also substantial noise). When I do vary HV to increase signal for dim specimens (channels), I use steps of 50 Volts: 550 V, 600 V, 650 V, 700 V, and rarely bother tryign higher (800 V, 850 V, 900 V) since the latter are quite noisy (Olympus is clueless about putting in 'better than averaging' options on acquisition - I could acquire time series with no averaging, but then would need to do clever things in MetaMorph [easy but somewhat tedious], learn MatLab programming (no thanks), learn Python, R, etc (no thanks), learn to program in Fiji ImageJ [no thanks and MetaMorph runs much aster). I also note that our Leica SP8 has two 2nd generatiion HyD detectors, peak quantum efficiency MAY be ~40% (or a little higher? ... 3rd gen is higher ... "4th gen HyD S is SiPM Silicon photomultiplier so for that detector "HyD" is just marketing slogan). The Leisca SP8 can "do" up to 16 line accumulation and 16 frame accumulation, so could do 256 accumulations, of photon counts. This would give superuior data to FV3000RS 16 line averaging,bt 256 accum would take a longtime ("time is money"). * Critical thinking - on your part, and don't believe everything you see in publications and marketing. * Review Biosearch Tech's STELLARIS RNA FISH galleries,https://stellarisgallery.biosearchtech.com --- their images are "maximum intensity projections" (MIPs) of Z-series so higher background than single image planes. Also they did not do 3D spatial deconvolution (slow in circa 2011 when gallery images data was produced). Especially useful (in my opinion) is their POLR2A image https://stellarisgallery.biosearchtech.com/Designready-Probe-Sets/i-gvtbKRH/A
Background into: pre-condition: low background by using 2D tissue culture cells, optically cleared, thin specimens (I claim thin sections are a physical type of "optical tissue clearing" -- thin 4um tissue sections in histology is partially to minimize out of focus background when using moderate to high numercial aperture (NA) objective lenses). Arjun Raj et al 2008 smFISH prefers 48 fluorophore-oligonucleotides (usually 20 bases each, single copy) on one RNA molecule, with 2 or more base gap (so minimum 1 kilobase RNA molecule -- 20 base also facilitates various math, such as 55% GC content =11 GC, 9 AT base pairs). This is partially because cells have RNA binding proteins (some sequence specific, some not) that 'cover up' some bindign sites, and/or double stranded RNA:RNA stems, so likely ~20 bind to any single RNA molecule (and may or not be random across the population of that RNA molecule]. Most RNA molecules are "sparsely distributed" in human tissue culture cells (more or less 2D flat on coverglass, usch as U2OS, HeLa, COS7 (which are African Green Monkey, but close enough), so rare that two RNA from same gene will be in same 3D location (aka point spread function, PSF, in micros-jargon). Also two, three or even several different RNAs unlikely to be "colocalized" in a single cell (unless crazy high expression like GAPDH ... one reason to use POLR2A, not GAPDH, as your labeling control). ** personal thanks to Marc Beal and Ron Cook for hosting me for a visit to Biosearch Technologies. Here is a photo of Ron Cook 9founder) and I with Kary Mullis' original DNA synthesizer -- designed, built and sold by Ron -- at Biosearch HQ (photo by Marc Beal):
https://works.bepress.com/gmcnamara/61 https://karymullis.com/pcr.shtml When I stumbled on PCR in the spring of 1983, I was trying to increase the demand for oligonucleotides, which before automation my laboratory had made by hand. Our new machine from my friend Ron Cook at Biosearch across the San Francisco bay had threatened job stability in the laboratory by doing what had taken us about three weeks to do, in eight hours—and it did it every eight hours, no breaks. My attempt succeeded. The demand went up by about a million and I didn't have to fire any of my fellow lab workers at Cetus.
|
Fluorophores -- more options listed at http://confocal.jhu.edu/mctips/multiplex
| Basic Five: DAPI + 4 Alexa Fluors | CyDyes | Brilliant Violets |
| DAPI | DAPI | BV421 |
| Alexa Fluor 488 | Alexa Fluor 488 | BV480 or 510 |
|
Alexa Fluor 568 * using R10/T90 can do Alexa Fluor 610-X see http://confocal.jhu.edu/mctips/af610-x (aND SEE BELOW) |
Cy3.5 (AF610-X) | BV570 |
| Alexa Fluor 647 | Cy5 | BV650 |
| Alexa Fluor 750 | Cy7 | BV785 |
| Alexa Fluor 700 (gm note AF700 cube arrived 12/2022: a user reported AF700 superior to Li-Cor IRDye700 for their smFISH probes - your benefit may differ) | Cy5.5 | |
|
Future -- since we lack funding (and get too little revenue) we would IDEALLY like to stop using the PentaCube --> AF568 "channel" and instead have THREE cubes optimized for each of: * AF546/AF555 * AF568 * AF610 Thiswould require about $5600 to $6000 assuming approximately same as our AF700 cube (~$1800). |
Quick summary of FISHscope:
Olympus IX 83 inverted microscope
Lumencor SPECTRA-III 360 light engine (fluorescence illumination, 8 single channels, we usually use at 25% power each to minimize photobleaching).
Semorock Penta LED filter cube (Penta ex, Penta dichroic, Penta em), Thorlabs R10/T90 beamsplitter, ThorLabs R03/T97 beamsplitter, CaliCube (EpiTechnologies - Tom DeMatteo)
- Semrock LED-DA/FI/TR/Cy5/Cy7-A
- excitation FF01-378/474/554/635/735
- emission FF01-432/515/595/681/809
- Beamsplitter FF409/493/573/652/759-Di01
- See https://searchlight.semrock.com/ for good graphing web site to compare our Penta filter cube with fluorophores.
The "RT" beamsplitters described at bottom. The CaliCube - developed by Tom DiMatteo -- enables visualizing the excitation path directly to the camera (mirror faces opposite of standard beamsplitter).
Alexa Fluor 700 cube (Semrock, installed 12/2022)
- Semrock AF700
- excitation 640nm laser from Lumencor SPECTRA III-360 (no exciter in cube, saved us some money, not needed and enables use of ANY excitation from the SPECTRA, i.e. 395nm for a BV)
- emission FF01-735/28-32
- Beamsplitter FF685-DI02-32X44-FX
- Cube: Olympus U-M710I Filter Cube (Semrock part OFX)
Sutter 10-3 emission filter wheel with 10 Semrock 32mm emission filters (full wheel) in deck 2 of IX83 (filters listed later - five match emission filter bands of Penta emission filter cube).
Hamamatsu ORCA-FLASH4.0LT sCMOS camera.
Olympus cellSens Dimensions image acquisition software with GPU deconvolution (50 cycles, "constrained iterative" -- on a Dell PC that is ok, not great. We have suggestions for a future "(near instant gratification" acquisition PC in McTips ).
- Experiment Manager [5D Experiment Acquisition] (GUI interface, may be faster acquisition than Process Manager)
- Process Manager -- we have acquired up to 20plex channels using this ... has:
channels
Z-series
"MIA" stage tiling and mark&find
- Well Plate Navigator
- Real-Time Panoramic Imaging
- EFI - Extended Focus Imaging (EFI) function
- HDR (High Dynamic Range) acquisition
- cellSens C.I. constrained iterative deconvolution (requires 2 or more channels, 3 or more Z-planes) using NVidia RTX 2080 Ti GPU
- For details on these cellSens Dimension 3.x "advanced features" details see https://www.olympus-lifescience.com/en/software/cellsens/5dexperiment-acquisition
Olympus objective lenses:
| Description | Mag | N.A. | working distance (mm) | Pixel size | GM Recommend Z-step |
| UPL SAPO | 4x | 0.16 | 13 | 1,620 nm | 5,000 nm |
| SP | 10x | 0.40 | 3.1 | 648 nm | 2,000 nm |
| UPL SAPO | 20x | 0.75 | 0.6 | 324 nm | 1,000 nm |
| UPL SAPO | 60x | 1.35 oil | 0.3 | 108 nm | 300 nm |
No 100x/1.4 NA or higher objective lens currently. We purchased the microscope on a "limited budget". We would love to have any or all (!!!) of:
X-line UPLXAPO100XO 100x/1.45 NA ... pixel size 65 nm
HR UPLAPO100XOHR 100x/1.50NA ... pixel size 65 nm
UAPON150XOTIRF 150x/1.45NA (https://www.olympus-lifescience.com/en/objectives/tirf-hr/) ... pixel size 43 nm
but (1) out of budget, (2) we like the pixel size on our camera of 108nm ("at specimen") becuse larger area (vs 100x or 150x lens) means more photons for ingle molecule RNA FISH, immunofluorescence, etc (no money for incubator either), and (3) if (when) we have more money, likely spend it on faster PC (recycling URL from above) future "(near instant gratification" acquisition PC in McTips ).
20210921: FISH reagent probe development "as a service" -
|
July 1, 2022: our NIH NIDDK P30 Center grant that funded the image core ended. As a consequence we are ending the "FISH reagents as a service". You can contact Prof. Bin Wu directly about the prospects of a collaboration with their lab on FISH reagents. We note that commercial reragents are readily available from Biosearch Technologies, see for example https://stellarisgallery.biosearchtech.com/Designready-Probe-Sets/i-gvtbKRH/A and Biotechne ACDBio RNAscope or equivalent from ThermoFisher. ** discontinued as a service New FISH Probe Synthesis (48 oligos) ... $200 (JHU internal price) * Our standard positive control probe set for human cells or tissues, human POLR2A, can be ordered as re-synthesis. Quantity per probe set is approximately 7-8 nmole per reaction. This is sufficient for approximately 500 slides or imaging dishes when using 12 mm diameter coverglass (12 mm round coverglass is 113 um^2). A 23 mm diameter is 415 mm^2, so 3.67x larger area, approximately 136 dishes. (ex. WPI FluoroDish, https://www.wpiinc.com/applications/neuroscience/fluorodish-cell-cultures ... WPI now also offers 10 mm diameter imaging area). A 20x20 mm coverglass is 400 sq mm, so ~150 coverglasses. We recommend imaging dishes for cells and for tissue sections (assuming the tissue section can be manueuvered into the dish imaging area). More information (JHU login required - users may also need to arrange iLab access through their dept/division and/or Research Dean's office): https://johnshopkins.corefacilities.org/sc/5148/ross-imaging-center/?tab=services
|
20210503M - Routine QA/QC capability (quality assurance / quality control) specimens
FISHscope and confocal (i.e. 60x/1.35 oil or 63x/1.4NA objective lens): The image core has ThermoFisher Molecular Probes TetraSpeck beads, Spherotech beads, including Ultra Rainbow, 5 intensities, 5.08 um diameter, mounted on slide (FPS-5057-UR5), same as 0.31 um diameter beads in suspension to mount on imaging dish etc (URFP-02-2), and Spherotech Fluorescent PMMA Particle Slide, “9plex”, 3.0 – 3.9 um diameter, mounted on slide (FPMAS-30M9). We do not use “PSF beads” because we prefer single molecule RNA FISH probe set cells on coverglass or imaging dish from Prof. Bin Wu’s lab, labeled with Bin’s standard fluorescent dyes (ex. Alexa Fluor 488, Cy3, ATTO 594, Alexa Fluor 647 or Cy5, Cy7; prefer probe set POLR2A, FISHscope acquire Olympus 60x/1.4NA, 108nm XY pixel size, 300nm Z-step size). We routinely use Olympus cellSens constrained iterative deconvolution (50 iterations, NVidia RTX 2080 Ti), and review the FISH probe set spots.
For our Horiba PTI fluorimeters, we have a set of six Starna Scientific fluorescent reference materials (6BF series polymer block references, https://www.starna.com/starna-6bf-series fluorophores in PMMA), of which Compound 610 and Rhodamine B are visible fluorescence in the range of our usual fluorimeter experiments, BCECF, GCaMP6s, mOrange2). We have not done so to date, we could uses the Starna blocks on our fluorescence microscopes.
GM usually starts new user training sessions with one of our Eosin fluorescent slides (from GM's collection, could obtain more from Nick Zachos' integrated physiology core's histology unit), which are bright, (relatively) photostable, have interesting structural details (skeletal muscle fibers details), surprisingly interesting spectral features (RBCs have red shifted fluorescence emission maximum, i.e. hemoglobin suppresses blue edge fluorescence), and some near infrared fluorescent features (dim, but easily observed in Cy7 channel on FISHscope, and somewhat on SP8 and FV3000RS).
20200831: A few throught about "future directions" (our focus now is on using what we have!).
- more cameras would enable simultaneous acquisition of additional fluorophores (ex: Brilliants tandems, Phitonex tandems), re: Hazen Babock's published 4cam (2018), has made 8cam (personal commun.), Babcock HP 2018 Multiplane and Spectrally-Resolved Single Molecule Localization Microscopy with Industrial Grade CMOS cameras. Scientific Reports 8: 1726. https://www.nature.com/articles/s41598-018-19981-z ... 4 (or 8) inexpensive CMOS camewras.
- Bigger field of view and/or back illuminated camera(s) ... i.e. 5.5 megapixel (8 megapixel) "bi" camera(s). Current front illuminated sCMOS cameras are 82% (modest width spectral peak) quantum efficeincy; back illuminated are 95% QE over a much wider range, and better QE in UV-blue and Red-NIR spectral range. That is, "total area under the curve" greatly favors "BI" over "FI". The only problem: some vendors charge a huge premium for their BI over FI.
- New PC: PCIe gen4 E-ATX motherboard, big power supply(s) [and line conditioner and UPS to protect), 2 NVidia RTX 3090 GPUs (20+ Teraflops each!!!! this ection written 1 day before NVidia launch date for Ampere architecture GPUs, PCIe gen 4 etc ... goa: "instant gratification" spatial deconvolution, that is deconvolve faster than acquisition time ... ideally "joint spatial deconvolution and spectral unmixing" [JSDSUN] ... re adam Hoppe 2008 Biophys J reported 10x improvement in SNR, but was slow at the time), NVMe gen4 SSDs (on motherboard and ASUS Hyper M.2 PCIe gen 4 NVMe array. Dual HD 4K monitors would be nice, though our current acquisition software (Olympus cellSens 3.0, spring 2020) is not really dual monitor friendly - ideally, most of the time: one monitor for acquisition controls, one monitor for images (i.e. one big picture, and thumbnails for each channel, image histogram, LUT controls, etc (the Leica LAS software for our Leica SP8 confocal microscope is closer to dual monitor friendly, but Leica has "missed the landing" on optimizing GUI controls). ... update May 2021 (hopefully the shortage of NVidia RTX 30x0 cards will be resolved before end of 2021): I suggest two RTX 3080 cards would outperform one RTX 3090 card (and about equivalent prices, nominally 2*$800 vs 1*1400) if the deconvolution (and joint spectral unmixing) software, data, and application needs were all optimized to take advantage of dual 3080s ... of course even more fun (on a server rack) could be four 3080 cards. Partly because two PCIE x16 (gen 4) slots enable faster data transfer (to and from ram/CPU) than one (I also assume AMD Ryzen ZEN3 CPU with either 64 lanes or "WX" 128 lanes, and compliant gen4 motherboard). I also note that current (spring 2021) RTX cards can use a larger memory apperture than the old default 256 MB.
- As usual, would also be great for next software release to have "print money" and "direct deposit (to my bank account)" buttons (for my login ... though the touch panel screen could have the buttons, not sure where any printed money would appear).
20200828F: We won Lumencor's Earthday 2020's grand prize: a $10,000 SOLA u-nIR 350-760nm white light fluorescence light engine. Eligibility rules included using a Lumencor lamp. Ours is Lumencor SPECTRA III-360 8 band light engine(detailed on this web page). Winning slide of 5 color fluorescence (2 color FISH, antibody, MS2, DAPI) was made by Lauren Blake, graduate student in Prof. Bin Wu's lab. Acquired (by GM) on FISHscope.
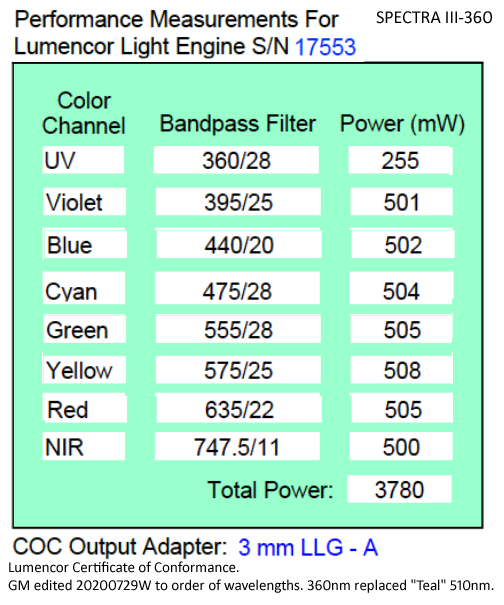
20200729W: I realized we have 555nm not 510nm LED in our SPECTRA-III-360 (360nm replaced usual 510nm). I added "Performance Measurements" table below (aka Lumencor Certificate of Conformance), may take some time before I replace all mentions of 510nm with 555nm. I discovered this because of testing with "RT" since 510nm would not be very useful with our Penta cube.
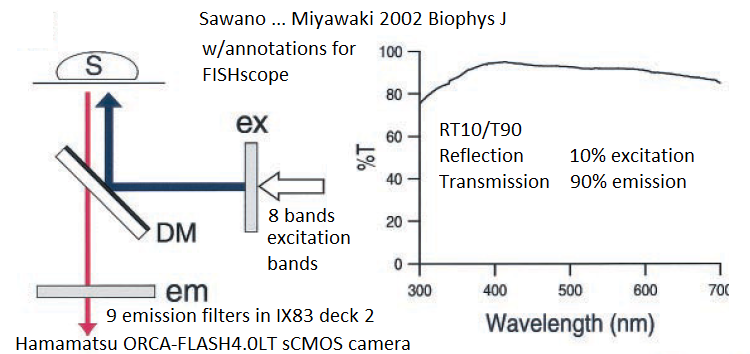
20200616Tue: new project: R10/T90 beamsplitter filter cube and "R03/T97" glass filter cube (re: Sawano ... Miyawaki 2002 Biophys J) to enable any combination of our Lumencor SPECTRA III-360 light engine and Sutter emission filter wheel with Semrock emission filters.Our preliminary results (6/2020) are each of these work, with somewhat longer exposure time on our Hamamatsu ORCA-FLASH4.0LT sCMOS camera (all controlled by Olympus cellSens). Advantage: not limited to "Penta" cube wavelengths, so we can use any of our Lumencor light engine's 8 exctiation bands. That is: "RT" provides a lot more flexibility than usual dichroics or multichroic beamsplitters.
20191111Mon" The JHU SOM Johns Hopkins Transcriptomics and Deep Sequencing Core Facility (Miller Research Bldg, room 351 and 360), http://www.microarray.jhmi.edu , is a good core to go to "upstream' of our FISHing capabilities. Use transcriptomics to identify the key RNAs, and then "Go FISH" with us.
20191107Thur: all is working. Lumencor SPECTRA III-360* installed, run through Olympus cellSens, "one click" for the up to five fluorescence channels (can be single plane or Z-series).
20191104Mon: Ross Imaging Center On iLab since November 4, 2019 ... restricted use (GM, Ana De La Cruz, maybe another Wu lab member(s), a very limited # G.I. Center members, future a few others), started 20190802Fri (August 2, 2019 after 'first light' 20190719F). We anticipate expanding use in the future (early 2020?), with priority being Ana De LaCruz experiments validating and using smFISH probe sets for G.I. experiments.
location: Ross Imaging Core (Ross Bldg 9th floor).
- Acknowledgement: All users are REQUIRED to include proper acknolwedgement of our funding sources. We suggest this text: The Ross Fluorescence Imaging Core FISHscope was funded by NIH NIDDK P30 DK089502 grant supplement to Prof. Mark Donowitz and Prof. Bin Wu, with JHU matching funding provided by Prof. Donowitz.
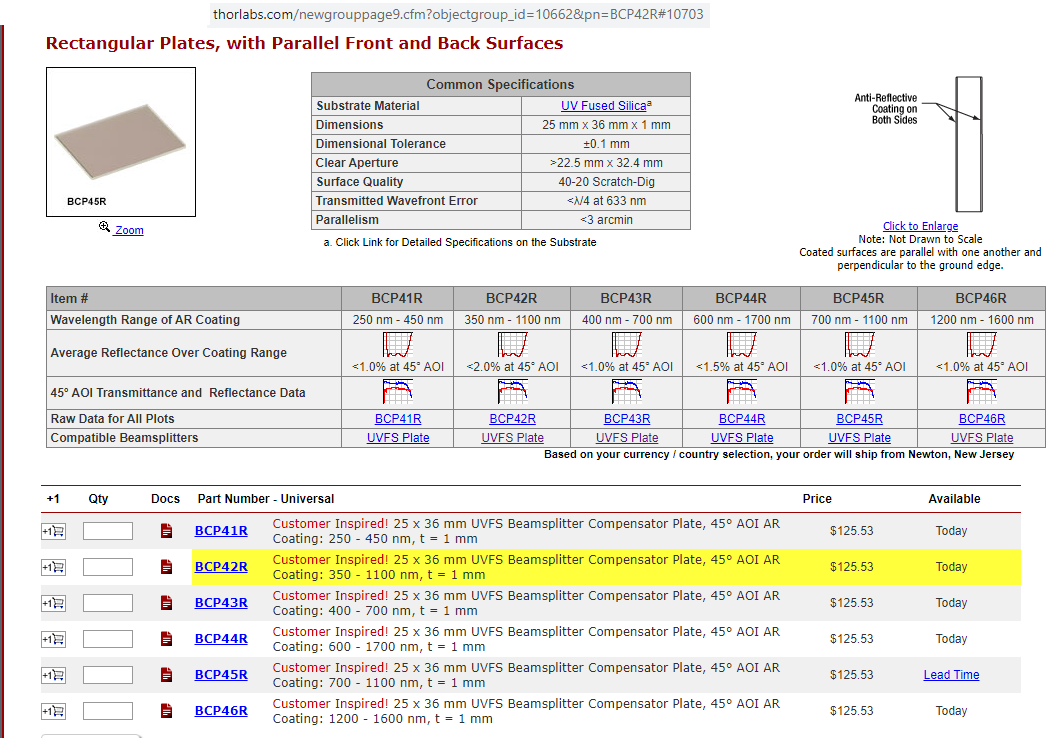
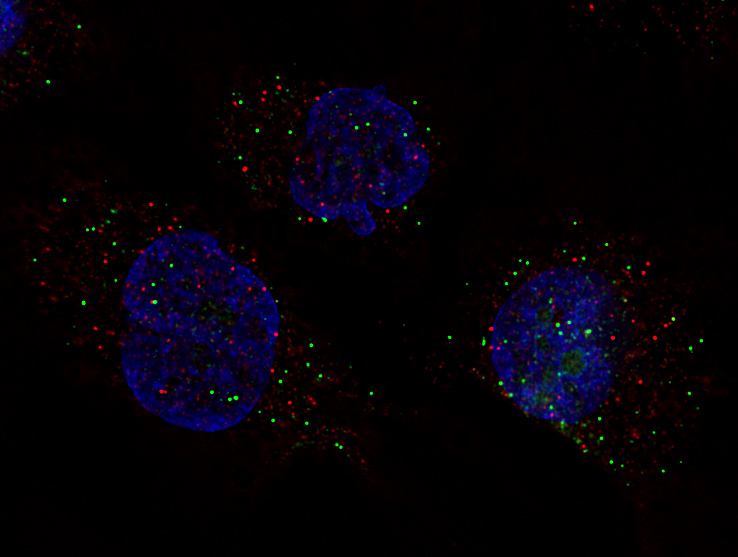
smFISH: one dot = one RNA molecule - below: POLR2A mRNA (green), EGFR mRNA (red), DAPI DNA counterstain (blue). See below for more details and full field of view.
- Olympus IX83 inverted microscope ... Olympus UPlanSApo 60x/1.35NA oil immersion objective lens (108 nm pixel size at camera). Also: Olympus 20x/0.75 NA lenses. Condenser 0.55 NA (brightfield).
- We note that our main objective lens, the 60x oil immersion lens, and our GPU Deconvolution module license (see next bullet) was purchased with our NIH shared instrumentation grant 1S10OD025244-01 (Prof. Brian O'Rourke and Prof. Mark Donowitz) for our Olympus FV3000RS confocal microscope. For logistic reasons, we suggest just acknolwedge our P30 supplement funding in your manuscripts.
- Olympus Cellsens Dimensions with GPU deconvolution (deconvolution license transferred from our Olympus FV3000RS confocal IX83 microscope NIH S10 grant purchase).
- Lumencor SPECTRA III-360* lamp, 8 excitation 'bands', 360 to 747 nm (initially using a demo SPECTRA X provided by John Gibas, Olympus) (Erich Zeiss, Iain Johnson, et al, Lumencor facilitated).
- Sutter ten position 32 mm filters emission wheel inside deck 2, Lambda 10-3 controller
- AVR Optics / Semrock filters: 9 (one empty position ... plan 7/2020 is to buy 10th emission filter to balance our wheel):
- Semrock Penta cube (ex + em + em filters in cube) LED-DA-FI-Cy3-Cy5-Cy7 + five emission filters in Sutter wheel. Semrock filters purchased through AVR Optics (Peter Brunt especially helpful).
- Semrock four (7/2020 soon to be five) additional emission filters to essentially "fill the gaps" between the Penta emission filters.
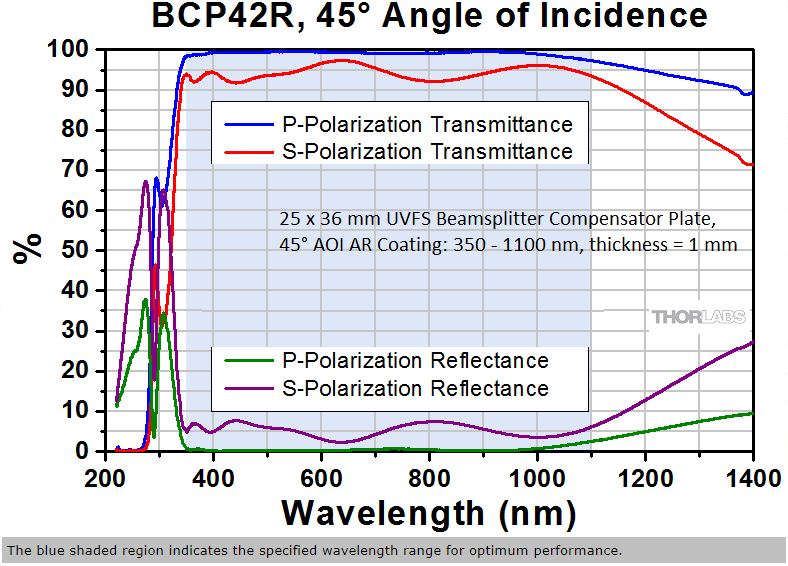
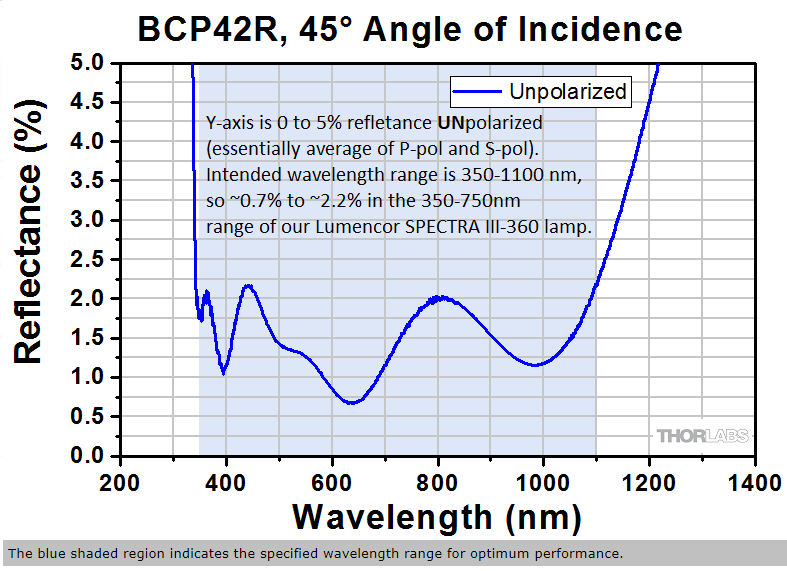
- Thorlabs "RT" cubes (added 06/2020) R10/T90 beamsplitter (Thorlabs BSN10R) and "R03/T97" glass (Thorlabs BCP42R, "compensator"). Enables excitation with ANY of our 8 excitation "bands" from our Lumencor light engine with ANY of our 9 emission filters. No need to change filter cubes (so less risk of image shifts than when using conventional DAPI, Green, Cy3, Cy5, Cy7, etc, filter cubes ... also faster). The "RT" beamsplitter is also less expensive than a standard dichroic or out Penta multichroic. More on "RT" near bottom (Torlabs product details and graphs).
- Hamamatsu FLASH4.0LT sCMOS camera (purchased through BBMicro / Hunt Optics) (Butch Granada, Hamamatsu, especially helpful).
- Our thanks to Kevin Murphy PhD, JHU Cardiology, and John Gibas, for leading getting our PC to Windows 10, Cellsens, all drivers, and improved performance.
- Our thanks to Ron at Olympus TAC (technical support) for microscope and Cellsens Dimensions assistance ("one click" Penta acquisitions).
- future ... we anticipate adding more fluorescence filter cube(s) and emission filters in the future [maybe not many given capabilities of our 6/2020 RT cubes) , to enable more applications (Brilliants [BUV, BV, BB, BYG], SuperBrights, S.ChuQDots, more ... i.e. BUV_longpass, BV_longpass, BB_longpass complete cubes --> full set of "adjacent emission bands" in Sutter emission filter wheel ... think "21plex" immunocytochemistry (I.C., aka "eye see") / immunofluorescence + smFISH ... Go "IC & FISH" ... pronounced "go ice fish(ing)"). For now focus is smFISH.
- No plans for environemental controls (for now): the FISHscope does not have a 37 C incubator, and we have no plans to purchase one. That said, in the future, we can contemplate - some day - moving the Tokai Hit incubator from our Andor X1 spinning disk confocal microscope, designated "Legacy status" August 2019, over to FISHscope.
smFISH: one dot = one RNA molecule - below: POLR2A mRNA (green), EGFR mRNA (red), DAPI DNA counterstain (blue).
Olympus UPlanSApo 60x/1.35 NA oil immersion lens, IX83 microscope, Lumencor SPECTRA X lamp (demo unit from Olympus), Semrock Penta cube (ex, dm, em filters in cube), single emission filters in Sutter 10-3 wheel (IX83 lower deck), Hamamatsu sCMOS camera (2048x2048 pixels), Olympus Cellsens constrained iterative deconvolution (NVidia GPU card accelaration), contrast adjustments and color combine with MetaMorph Imaging System. 108nm pixel size, ~200x200 um field of view. Note: pixel scale 1/4 and saved as JPEG quality 9 (max is quaality 12) due to limitations of our web site host.
Image credit: Ana De La Cruz, JHU SOM.
![]()
How many plex do you want to image?
Note: Olympus cellSens (as of 6/2020) can list 40 acquisition settings in Process Manager.
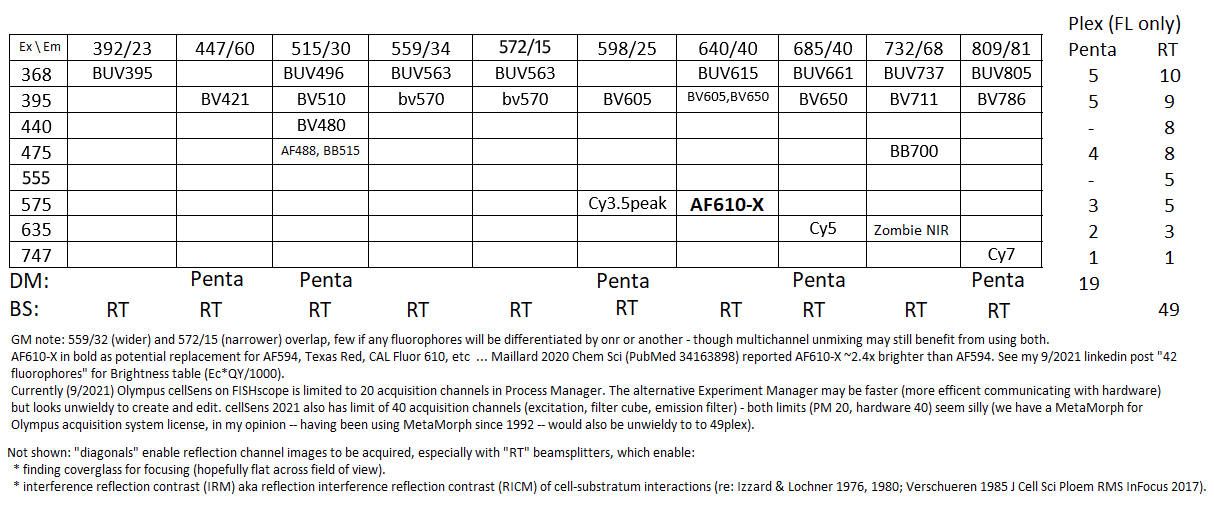
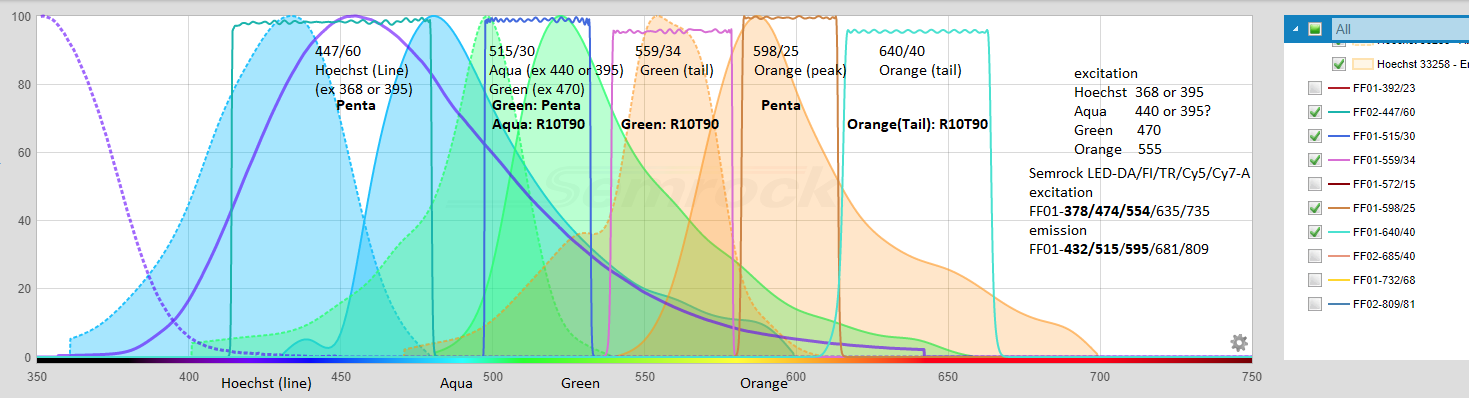
McNamara 20200624W FISHscope 8 excitation 9 emission bands – table (note; we are in process of ordering filter #10 to balance the wheel and add yet another eission band option).
Lumencor light engine (368 .. 747 nm) --> RT beamspliiter cube --> specimen --> Semrock filters in Sutter filter wheel 392 .. 809 nm) --> Hamamatsu sCMOS camera
* GM note; "510" (left column) should be "555" (will fix table), also now have 572/15 emission filter installed (replaces "Empty").
GM notes: reflection is useful for
(i) find the coverglass-mounting medium surface,
(ii) flatness of the coverglass [or lack thereof .. and whether the specimen should be re-positions to make as flat as possible],
(iii) interference reflection contrast microscopy (IRM, aka RICM … optional dual or triple wavelength quantitative IRM, qIRM). In principle, for fluorescence, could acquire every emission band longer than excitation (which are labeled as ‘reflection’ here): 9 + 8 + 7 + 7 + 6 + 5 + 3 + 1 = 50 images. If each image is 1 second exposure time (data transfer is ~10 ms each, so 500 ms total; emission filter wheel moves could be minimized by stepping only between excitation wavelengths, so 8 moves in ~50 ms each, so ~400 ms; more conservative in case of short wavelength induced photobleaching is to excite longer à shorter excitations, so ~50 moves * 50 ms = 2500 ms) could acquire all in 50.5 seconds, so under 1 minute (in practice, some may be optimal at longer exposure). Note: our Lumencor light engine has “plenty of power” in each excitation band, and our “RT cubes” work at every wavelength (reflection/transmission, R10/T90, R03/R97). Excellent IRM (and qIRM) review: Barr Bunnell 2009 Interference reflection microscopy. Curr Protoc Cell Biol, Unit 4.23. PMID: 20013754. More on IRM below (near bottom).
20210921U: Ex \ Em table (revision) with plex and comments. One reason for adding this version is finding that Alexa Fluor 610-X is ~2.4x brighter than Alexa Fluor 594 (Maillard et al 2020 Chem Sci).
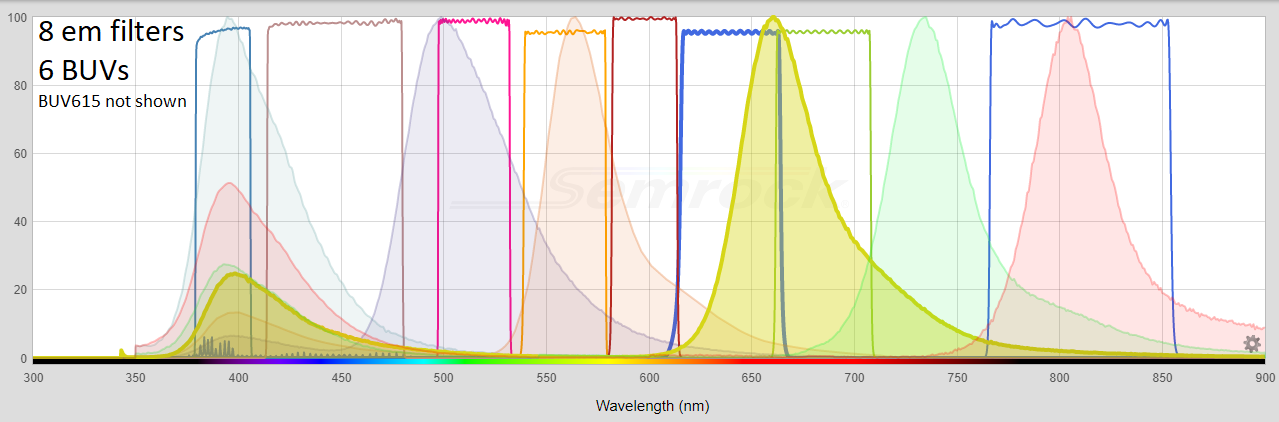
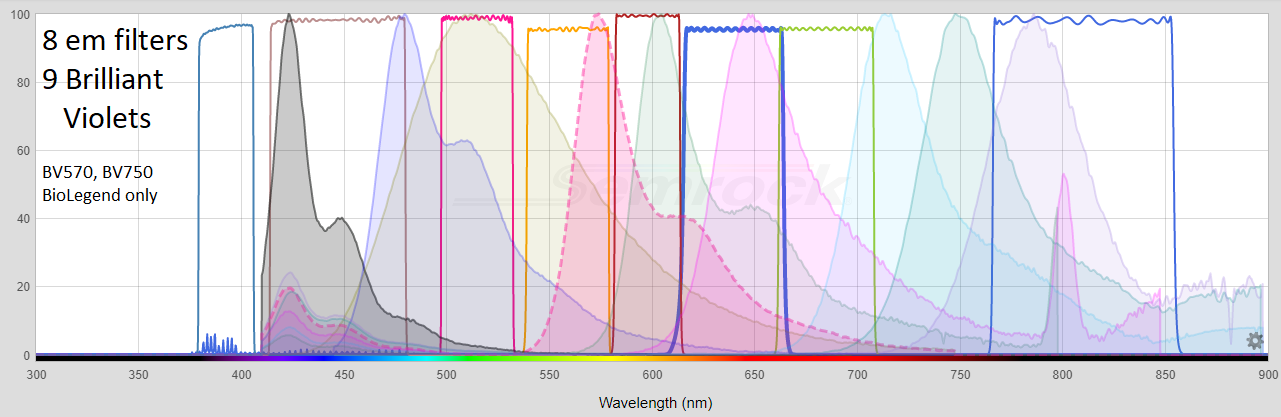
Below: table 20200623Tue (older version of table, including Reflection), with RT10/90 (R10/T90) and "RT03/97" (R03/T97) beamsplitter cubes (R-Reflection, T=Transmission, numbers are percent of light; see above for more info on these cubes). Also tabulated are Brilliant Ultraviolets (BUVs) and Brilliant Violets
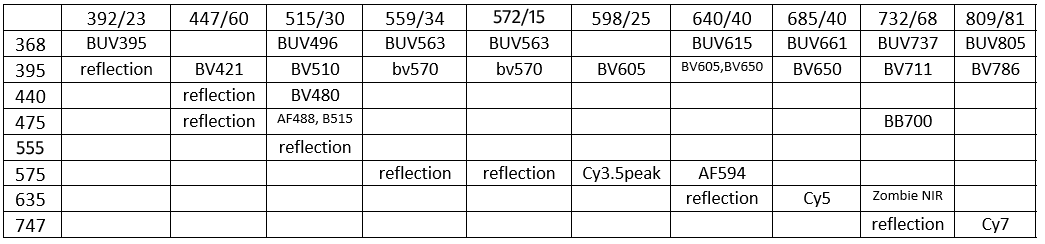
(BVs). Two BV's available only from BioLegend (BV570 and BV750), which licensed BV's from Sirigen before BD Biosciences acquired Sirigen.
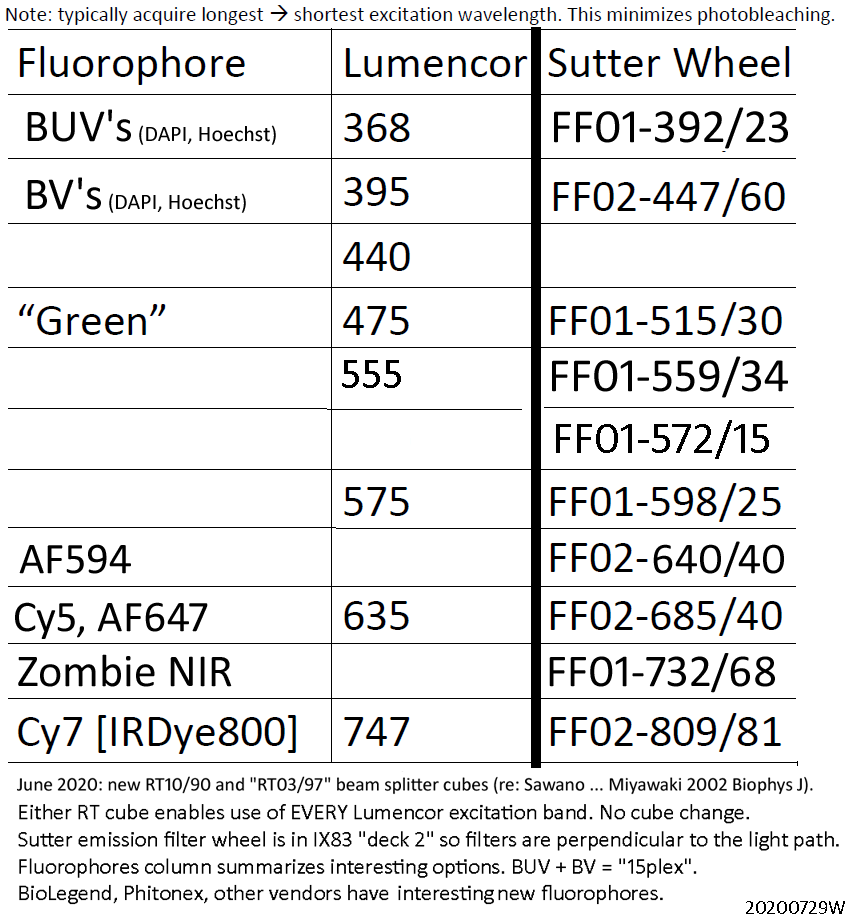
Below: "Go FISH" Prof. Bin Wu currently likes: Cy3, Alexa Fluor 594, Cy5, Cy7 for single molecule RNA FISH probe sets (alternatives: Atto 594 for AF594 (GM: see also AF610-X), AF750 for Cy7, not shown). Also shown BUV395 and BV421. All accessible using either "RT"cube and one or more emission filters (and enabled by having a nice camera: Hamamatsu ORCA-FLASH4.0LT).
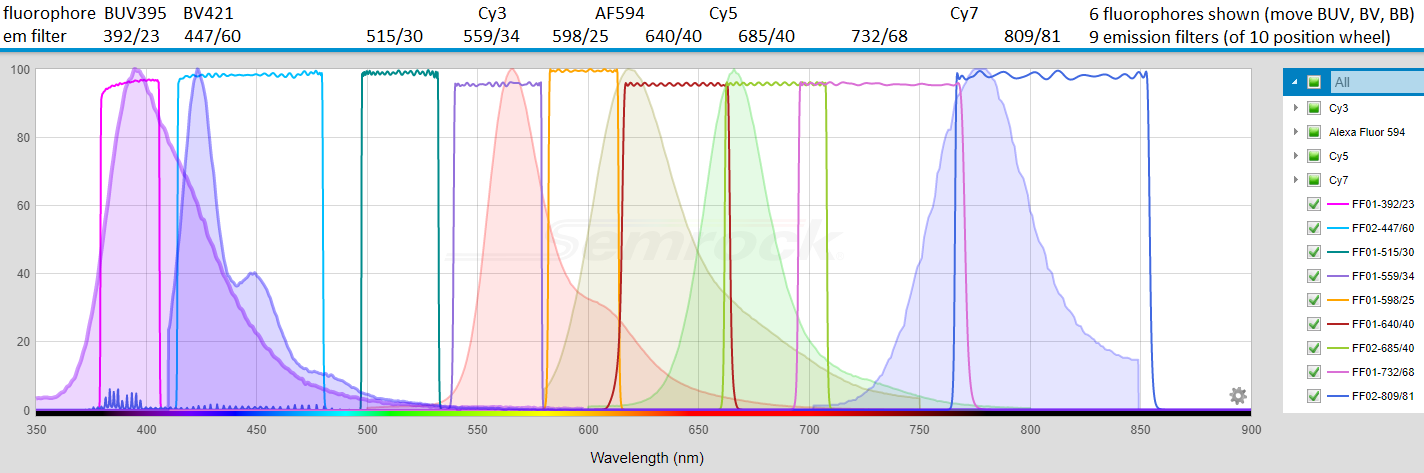
Below: BUVs and BVs with respect to FISHscope (640/40 em and 732/68 on order 6/2020).
All Emission filters, from https://searchlight.semrock.com - Penta cube shown below.
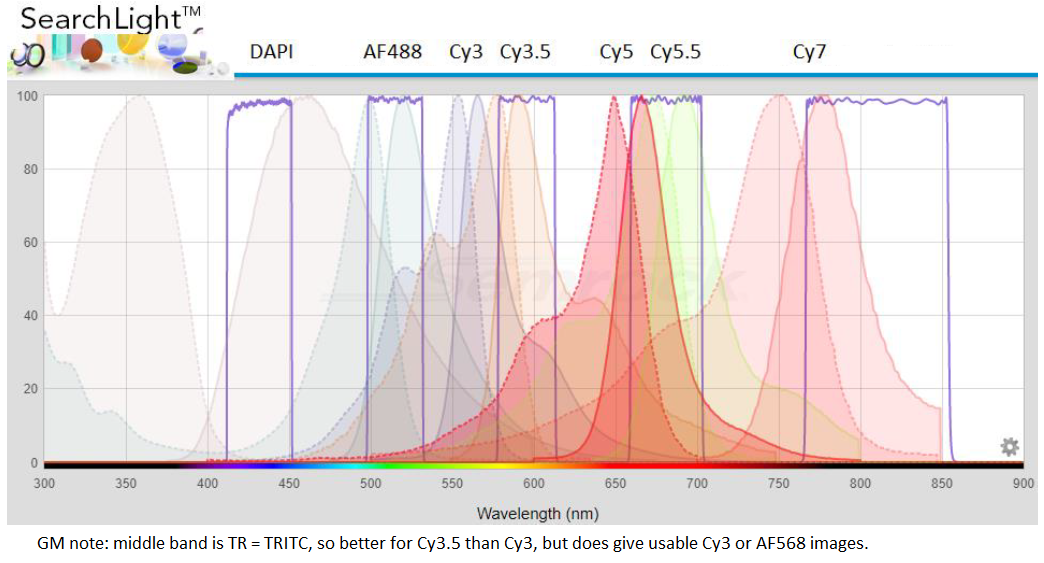
20220421H: below is graph from Semrock Searchlight + GM annotations for Hoechst (or DAPI), Spectrum Aqua, Spectrum Green, Spectrum Orange - typical for DNA FISH. I note on FISHscope the R10T90 beamsplitter cube is "U-FUN" on the Olympus IX83 TFT touch panel (acquisiton is automated in Olympus cellSens where it is labeled ufun).
**
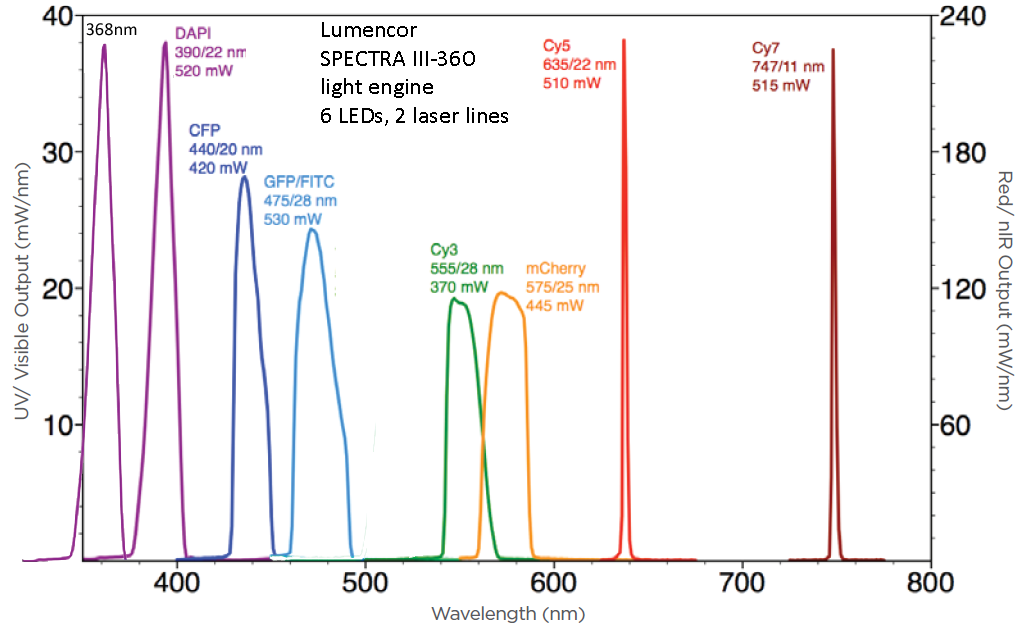
Power Measurements (Lumencor "Certificate of Conformance") with "color", wavelength (and LED emission filter; last two are laser lines) and power measured at factory.
***
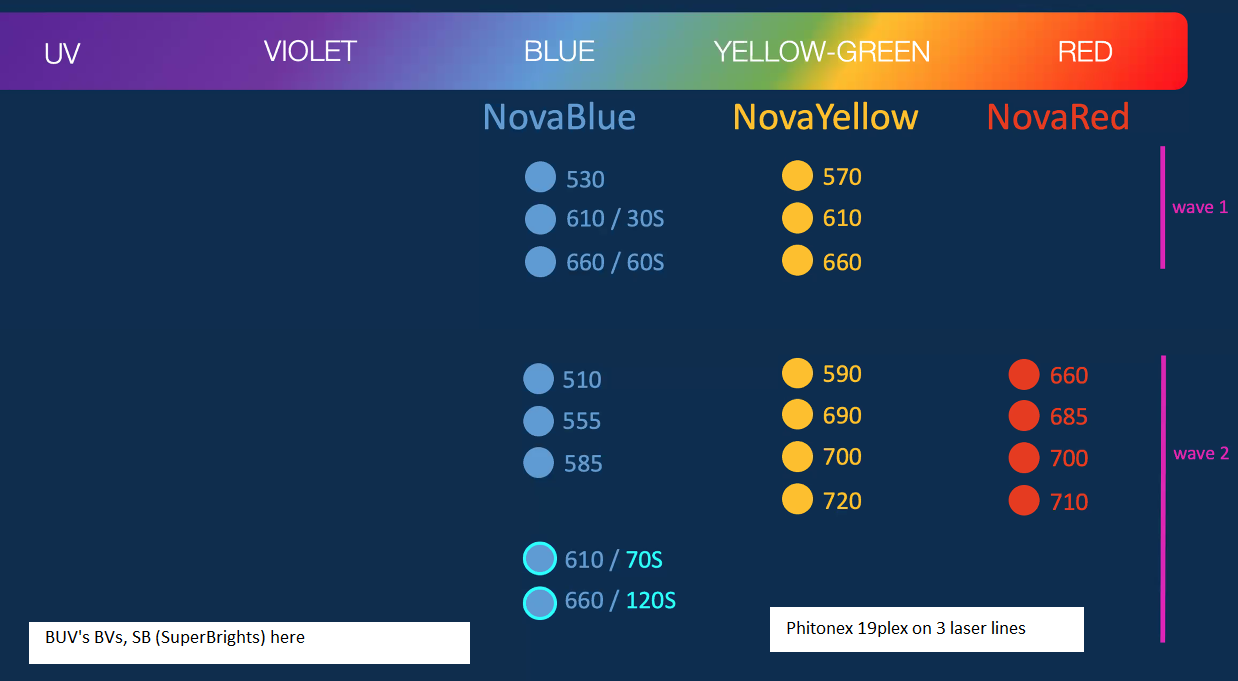
News 20200624W: Phitonex 19plex Nova's ... can be used with Brilliants BV, BUV, SBs (SuperBrights) ... Phitonex is targeting flow cytometry market, looks promising for fluorescence microscopy.
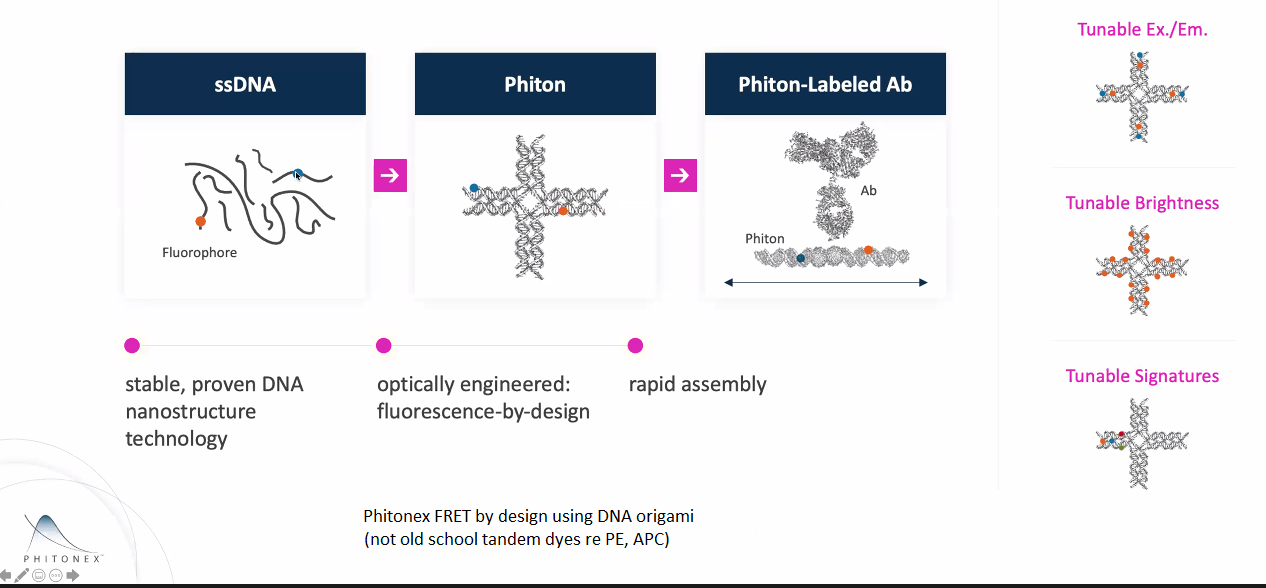
Phiton DNA origami ... fluorescent dye(s) on or in the DNA origami ... built from oligonucleotides (see Phitonex web site and white papers for academic references). The Phiton 150 kDa "cruciform structure" is 20nm linear, but folded into a 3D "cup like structure". Upshot is similar in molecular weight (150 kDa) and dimensions to an antibody. Normal use would be direct label on a primary antibody (and probably defined 1:1 or 1:2 antibody:phitons). Would be possible to conjugate to Fab (~50 kDa) or nanobody (~12 kDa). Note: we do not have any financial interest in Phitonex, just intrigued by potential for multiplexing. See also Ron Vale's 2020 Nature Methods "FluoroCube" (6 nm cube, published 6 identical dyes on 6 of the 8 corners, one corner open for conjugation ... I don't understand why they did not put dye on 7 of the 8 corners).
************************************************************************
A key accessory to enable additional uses is the LCI 24 well plate optogenetic device, funded with the P30 supplement + Mark Donowitz matching funds:

|
20200713Mon - more information on our RT Beamsplitters project. Goal: eliminate need for dichroic (2-color) or multichroic beamsplitters in our microscope. Our initial filter cube was a "Penta" cube (ex+dm+em each "penta") + 5 matched emission filters in wheel from Semrock (U.S. distributor AVR Optics). Works fine for the "penta" (5plex) designed for: DAPI / FITC (green) / Cy3 / Cy5 (Alexa Fluor 647) / Cy7. Can be used for other fluorophores, such as some Brilliant Violets (395nm excitation, various emission), but not all. Concept - use a piece of glass -- from Sawano ... Miyawaki 2002 Biophysical Journal (see Sawano ... Miyawaki 2002 Biophys J open access ... figure 1 modified by GM)
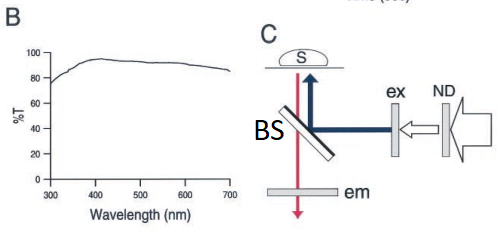
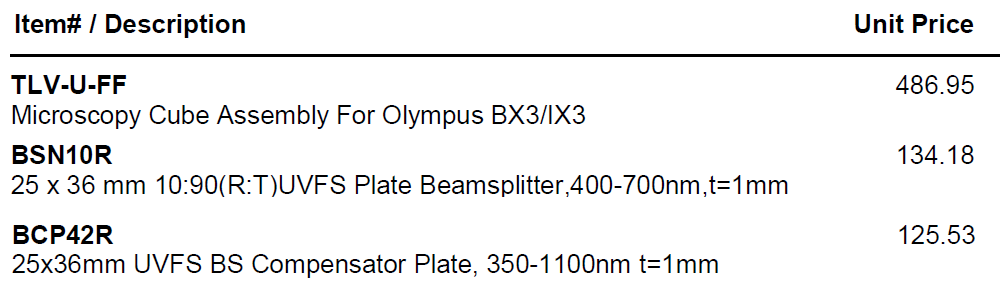
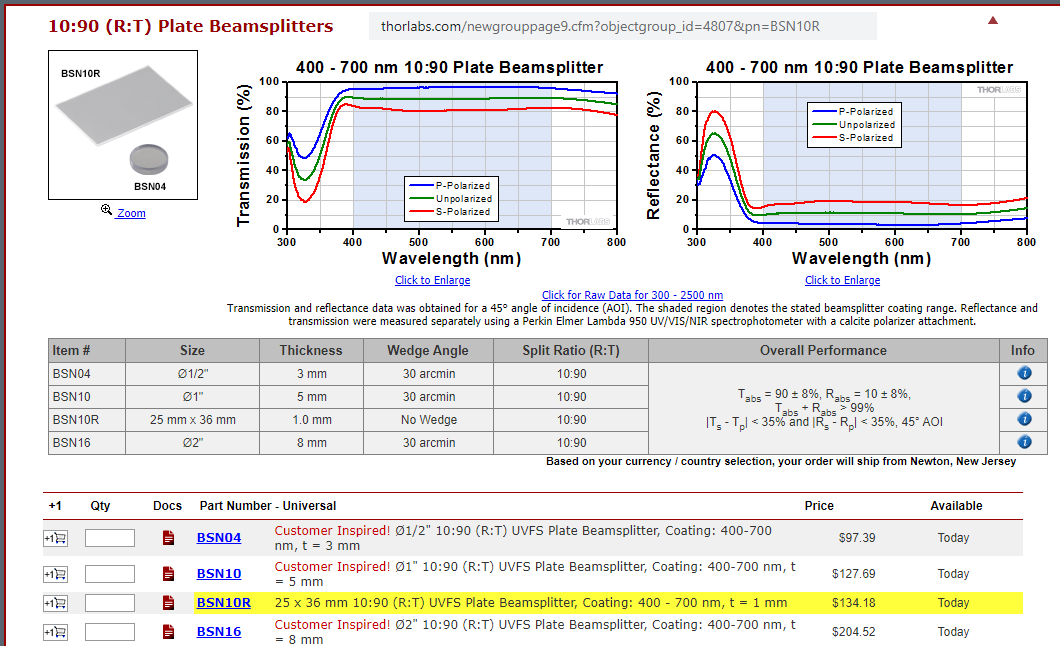
Doable: because we have a powerful lamp (lumencor SPECTRA III-360) and emission filter wheel in the microscope (so safet to use eyepiece or camera) and Hamamatsu ORCA-FLASH4.0LT sCMOS camera. Implementations: 1. R10/T90 = reflection 10% and transmission 90%. Thorlabs BSN10R beamsplitter. 2. R03/T97 = Reflection ~3% and transmission ~97% Thorlabs BCP42R compensator (maybe more like 2% reflectance).I find ~4x longer exposure for this compared to R10/T90. Each in an Olympus IX83 filter cube (these two from Thorlabs, TLV-U-FF filter cube) -- only the beasmplitter, no exciter, no emission filter in cubes, since our microscope has Lumencor lamp (8 single exciter 'bands') and 10 emission filters in Sutter emission wheel (deck 2 of IX83 microscope).
Thorlabs web site has prices for these items and most other products. Thorlabs beamsplitter specifications.
BSN10R
BCP42R compensator
BCP42R reflectance (0 to 10% depending on polarization state and wavelength) and transmittance (90 to ~98%). We care about;
BCP42R ... UNpolarized reflectance (excitation light) - note wavelength range on graph is 200-1400nm, the FISHscope wavelength range is 360-880-nm (368nm excitation to right end emission of 809/81 emission filter is 850nm).
7/2020 initial results: For our Eosin tissue section slide, I obtained equivalent intensities with 555nm excitation and 598/25 emission filter in wheel for: * Penta cube ... 150ms * R10T90 ... 1000ms * R03T97 ... 4000ms using same Lumencor 555nm intensity level (25%). I could have adjusted lamp level (increase for RT10T90, increase more for R03T97), but did not since 4 second exposure not a big deal time size nor dark current noise on our camera.
|
|
IRM = interference reflection contrast microscopy RICM = reflection interference contrast microscopy From above: interference reflection contrast microscopy (IRM, aka RICM … optional dual or triple wavelength quantitative IRM, qIRM). In principle, for fluorescence, could acquire every emission band longer than excitation (which are labeled as ‘reflection’ here): 9 + 8 + 7 + 7 + 6 + 5 + 3 + 1 = 50 images. If each image is 1 second exposure time (data transfer is ~10 ms each, so 500 ms total; emission filter wheel moves could be minimized by stepping only between excitation wavelengths, so 8 moves in ~50 ms each, so ~400 ms; more conservative in case of short wavelength induced photobleaching is to excite longer à shorter excitations, so ~50 moves * 50 ms = 2500 ms) could acquire all in 50.5 seconds, so under 1 minute (in practice, some may be optimal at longer exposure). Note: our Lumencor light engine has “plenty of power” in each excitation band, and our “RT cubes” work at every wavelength (reflection/transmission, R10/T90, R03/R97). Excellent IRM (and qIRM) review: Barr Bunnell 2009 Interference reflection microscopy. Curr Protoc Cell Biol, Unit 4.23. PMID: 20013754. More on IRM below (near bottom). References (most can be found through PubMed https://pubmed.ncbi.nlm.nih.gov/ to the journal article online): Abercrombie Dunn 1975 Adhesions ... interference reflection microscopy. Exp Cell Res 92: 57-62. Barr Bunnell 2009 interference reflection microscopy. Curr Protoc Cell Biology 45:4.23.1-4.23.19. DOI: 10.1002/0471143030.cb0423s45 Curtis 1964 The mechanism of adhesion of cells to glass - a study by interference reflection microscopy Nong 2021 bioRxiv - integrated multi-wavelength SCATTIRSTORM microscope combining TIRFM and IRM modalities for imaging cellulases and other processive enzymes https://doi.org/10.1101/2021.02.25.432940 Ploem Prins 2017 Reflection-contrast microscope - Review. RMS In Focus 47: 38-56. (with lots of great refferences). Ploem 2021 http://ploem-reflection-contrast-microscopy.com Schilling ... Sackmann 2004 Absolute interfacial distance measurements by dual-wavelength reflection interference contrast microscopy. Phys Rev E 69: 021901. Verschueren 1985 interference reflection microscopy in cell biology: methodology and applications. J Cell Sci 75: 279-301. *** Help! I need You -- or someone you know -- aka free labor with computer programming andsoftware development skills I would like someone (you? if you are a student; one of your mentees if you are a PI/mentor) - or anyone could just "DIY" and let me know about it -- to develop an image processing 'app' -- probably simplest as a Fiji ImageJ plugin ( https://imagej.net/Fiji/Downloads -- see klinks at far right of page for programming stuff ) to implement Schilling ... Sackmann 2004 dual wavelength RICM (quantitative IRM) and even better, triple wavelength RICM. That is, you do the 99% perspiration, and I provide the 1% inspiuration. I am happy to comment on (aka 'edit)' your manuscript -- you = first author (or co-first author if you are plural), and I be acknowledged or a coauthor, as you deem appropriate (and assuming I do any work, like helping write the manuscript and acquire and provide training). I can provide images from FISHscope and probably other microscopes, i.e. our Olympus FV3000RS confocal and/or Leica SP8 confocal microscopes.
|
Go FISH! - (bulk) RNAseq vs smFISH
We found that increasing MYC levels increased the expression of most genes (Figure 3C). We picked four genes— RPAP3, RAB7A, KPNB1, and MYH9 — for more detailed analysis. We note there is a discrepancy in rank order of expression when comparing RNA-seq and smFISH for RAB7A and KPNB1. All subsequent experiments utilize smFISH, which we found to be a more robust measurement of expression levels.
MYC amplifies gene expression through global changes in transcription factor dynamics.
Patange S, Ball DA, Wan Y, Karpova TS, Girvan M, Levens D, Larson DR.
Cell Rep. 2022 Jan 25;38(4):110292. doi: 10.1016/j.celrep.2021.110292. PMID: 35081348
..
Reserve Equipment