McTips_2024
McTips_2024 ... newest posts at top
George McNamara, PhD, gmcnamara@jhmi.edu
... unfortunately my main McTips page reached some content limit (we don't pay much per year to the web hosting company), so stuck using subfolder.
|
https://www.linkedin.com/posts/kiryl-piatkevich-6b83b341_the-mscarlet-family-of-fluorescent-proteins-activity-7269516992334692355-Kl1f The mScarlet family of fluorescent proteins keeps growing! Meet mScarlet3-H (aka mYongHong), an ultrastable RFP ideal for many advanced microscopy applications. Plasmids are now available via WeKwikGene! https://lnkd.in/gjE3Vmzs https://wekwikgene.wllsb.edu.cn/plasmids/0000716 note: bottom of page has .DNA file (mostly text); mScarlet3-H kozak sequence is ~1000 (i.e. look for AUG start codon close to 1000 if using a text program). Plasmid map on web page. |
||||||
|
interesting new deconvolution software https://www.mbfbioscience.com/products/neurodeblur At the heart of NeuroDeblur are state-of-the-art algorithms for synthetic PSF generation and adaptive background removal. Our extensively validated synthetic PSFs enable excellent deconvolution results for both light-sheet and confocal data, eliminating the need for PSF measurements. The sophisticated rolling-ball background removal preserves fine details while removing inhomogeneous background, resulting in strikingly clear images. |
||||||
|
Evident Scientific new Silicone Gel Pad(s) as alternative to their Silicone oil for our Olympus FV3000RS confocal microscope linkedin thread (part of thread -- *** separators) https://www.linkedin.com/feed/update/urn%3Ali%3Aactivity%3A7267659767873978368/?comment Why did Evident Scientific innovate the LUPLAPO 25XS/0.85NA long working distance (2mm) Silicone Gel Pad objective? The solid Gel Pad offers the ease of using a dry objective with the refractive index of silicone oil. *** GM: Hi Craig, how is the silicone gel pad cleaned? Does it work with all the S.I. lenses? Happy Thanksgiving, George *** CR: They are disposable. The spec is 30 cycles of a 96 well plate but the objective has just been launched so excited to see how customers like it. |
||||||
|
Cute trick: endoscope camera to inspect settings on objective lens correction collar (I wonder is a simple web cam would work) ... also known as Borescope camera (typically with light, some models have small viewer), example https://www.amazon.com/Articulating-Adjustable-Waterproof-Automotive-Semi-Rigid-dp-B0CBCBSYDG/dp/B0CBCBSYDG * amazon also has various USB microscopes, with basic or slighly fancier stands, optional lighting, viewer.
Microscopy and Imaging Center Texas A&M University |
|
20241118M Cool relative Ca++ imaging tip - also any fluorescence intensity quantitation from confocal listserv Hi Gabor, Question was: Dear All, ** GM notes: * classically, Ca++ ion concentration was measured with Fura-2, using 340/380 nm excitation ratio and "wide green" detection (i.e. 500-550nm). Not ideal because of UV light inducing phototoxicity (kill cells) and photobleaching. Worked nicely on widefield microscopes with the ratio correcting for thickness of cell(s) at different pixels (voxels). * GCaMP6 and other fluorescent protein based Ca++ ion biosensors have mostly superceded Fura-2, done with cofnocal microscope to optically section cells (though thickness of cell vs NA of objective lens need to be checked). * Various other ratio tricks are available, for example using visible wavelengths (most modern microscopes lack 340 and 380nm excitation), could use GCaMP vs "inert" red fluorescent protein (though all FPs are pH sensitive, and would ideally want ideal pKa's). Another option is Fluo-3 or Fluo-4 (each green sensitive) and Fura-Red (red; inverse intensity wrt Ca++ ion concentration, expanding ratio). |
|
GM notes: CCD = Charge couple device (in 2024, most modern research microscopes use sCMOS = scentific CMOS [see online acronym] or point scanning confocal microscope (latter enables zoom and field of view to decide pixel size). PSF = point spread function. FWHM = full width at half maximum. confocal listserv URL https://lists.umn.edu/cgi-bin/wa?SUBED1=confocalmicroscopy&A=1 ------------ Guy Hagen - ------------- Confocal listserv Mon, 18 Nov 2024 at 11:46, Steffen Dietzel <lists@sdietzel.de> wrote: Look at pages 130--133, Section 5.9: Pixel sampling. He states that 2.3 is the FWHM in pixels of the Gaussian approximation to the PSF. {GM note: chp 5 is available from CUP URL below). Howell text (see the book or eBook - chp5 online through JHU -- for more text and several figures): Confocal listserv question that led to Steffen D post citing the Howell text:
*** GM's take: Biological objects are rarely a single point, so rarely a single point spread function (PSF) data. Also rarely sine Waves, re; Nyquist-Shannon sampling theorem. Enought pixels need to be acquired to resolve the object of interest -- that is resolve details in one object or twll if two or more objects. Some issues: * is the object or objects in focus? ... if not, maybe acquire 3D Z-series with high enough sampling (small Z-step interval) (and GPU deconvolution). * is the object(s) precisely aligned with the detector's horizontal or vertical (doable by rotating the object, though not trivial, or rotating the detector [not goign to get my permission to do this, since our cameras are aligned to the XY stage, or rotate the scan direction of a confocal microscope). * is the object(s) orientation(s) random or along the diagonal ... if the latter, than sqrt(2) = 1.4142 needs to ab accounted for. * is resolving really needed? If using 50nm pixels (projection at the specimen), each pixel gets 25% of the photons of 100nm pixel size (see Howell text and figures for nice example of of "large pixel sampling" for PSF centered on a pixel vs at the intersection of four pixels. For single molecule localization microscopy, the goal is centroid calculation, assuming sparse fluorophores "on", so most SMLM microscope data acquired with 'somewhat large' pixel size. My thinking - if want to resolve and have the hardware, is to do this math: 2.355 * sqrt(2)= 3.33, and hence resolution: dxy(widefield) = 0.61 * wavelength / NA dxy (confocal 1.0 Airy unit) = 0.51 * wavelength / NA dxy (confocal 0.66 Airy unit) = 0.9 * 0.51 * wavelength / NA ... 0.9 from a Zeiss PDF; see also Jeff Reece (NIH) "confocal sweet spot (range)". dxy can be improved by ~10% by deconvolution (Paul Goodwin chapter - was at Applied Precision, re: DeltaVision, OMX). dxy(GPU deconvolution) = 0.9 * dxy(microscope mode above). GM pixel size (if need to resolve) = dxy(deconv) / 3.33 (in practice, I may divide by 3.0 or 3.33 or 3.5 ... but ultimately usually the limit is either optics available or project need ... or photobleaching ... or microscope stability (latter can be mitigated by combining vibration isolation table and "fiducial markers"). Wavelength matters: BUV395 was 395nm emission maximum (peak), so ~20% smaller PSF than Alexa Fluor 488, AausFP1 or EGFP (~520nm), in turn far smaller than Alexa Fluor 647 (em max ~660nm). On the other hand, brightness and photobleaching matter. If resolution is massively critical, reflection confocal microscopy, same wavelength for excitation and emission, with a reflection = scattering object would be ideal. NanoGold (www.nanprobes.com) is available in 1.4nm and 5nm diameter but probably scatter so little, would be hard to image (I've never seen a publication; nor active users). Collodal Gold (Au nanoparticles = AuNPs) or silver enhanced nanogold, either ~50nm, can be imaged by confocal. Maybe more practical: use Expansion Microscopy (ExM), for example "20ExM" (11/2024 Ed Boyden -- next section). |
|
20241111M update - 20xExM Single-shot 20-fold expansion microscopy ==>The ideal microscope for imaging 20xExM is likely to be FISHscope (with GPU deconvolution) since this research grade widefield fluorescence microscope is much faster imagng than confocal microscopes. Our current high resolution objective lens is our 60x/1.35NA, 108nm XY pixel size, recommend Z-step 300nm, working distance 300 um (so maximum 15um "original size" if use full 20x expansion). Working distance of most Evident Scientific (ex-Olympus) high resolution (X-Line and HR) objective lenses have smaller working distances (i.e. 1.45 and 1.50 objective lenses) and the 20x vs (say) 10x expansion doubles resolution anyway. A couple of potential tweaks to image better and brighter and less photobleaching (many also mentioned elsewhere in this and other McTips -- and often applicable tostandard fluorescence microscopy):
With 20-fold expansion, researchers can get down to a resolution of about 20 nanometers, using a conventional light microscope. This allows them see cell structures like microtubules and mitochondria, as well as clusters of proteins. In the new study, the researchers set out to perform 20-fold expansion with only a single step. This meant that they had to find a gel that was both extremely absorbent and mechanically stable, so that it wouldn't fall apart when expanded 20-fold. |
|
20241108F - FISHscope cell details 20x 0.75NA vs 60x 1.35NA raw vs GPU deconvolution - details can matter. Note: no images in this section - the acquired images are for the user and their lab, not for my essay. Yesterday I trained a new user on FISHscope http://confocal.jhu.edu/current-equipment/fishscope Description Mag N.A. working distance (mm) Pixel size GM Recommend Z-step UPL SAPO 20x 0.75 air 0.6 324 nm 1,000 nm or 500nm Sub-confluent thin cells (~4 um thickness?) growing as well separated colonies on a coverglass. The background fluorescence was much higher in the dense middle of large coonies compared to colony edges and small colonies. Several observations on my part: * unknown coverglass thckness - for specimens with potential high resokution imaging, buy and use #1.5H (H = High Precision, 170 +/- 5 um) thickness coverglass (and double check that 2 coverglasses are not stuck together) -- see box below (20241014M). * huge variation in colony sizes: this type of variability potentially affects both image quality and the biology they are studying.
* 60x oil lens, 25% 395nm LED power: DAPI not only photobleached but photonverted to green fluorescence - nuclei became bright green in the "Alexa Fluor 488" channel. Mitigation:
* We initially trained on 20x, single plane. Not much subcellular detail. * 60x single plane raw images were more "somewhat more" informative. * 60x Z-series raw data (22 or 30 planes, 300nm Z-spacing) better, but blurry (because 60x/1.35NA lens has ~600nm in-focus, but if cell is 3000nm thick, then ~2400nm "not quite" focused). * 60x Z-series GPU deconvolution (Process menu -> Deconvolution -> Constrained Iterative, 25 iterations) was was much more informative than 20x single plane or 60x raw. I note that the "C.I." algorithm on FISHscope requires Z-series, though maybe not as thick as we did (though maybe would have been even better if we did say 12um instead of 9um = 9000 nm of 30 planes 300nm Z-steps). Key item: the deconvolved Z-series best focus planes clearly showed sub-cellular immunofluorescence details, such as endosomes and endoplasmic reticulum, not readily recognized in raw data. * Junk on coverglass: there were a lot of small "objects" (puncta?) on the coverglass - readily apparent in the GPU deconvolved Z-series (Z-panes at the coverglass). I suspect "antibody aggregates" and recommend:
* huge variation in colony sizes - a decade ago at UMiami I coauthored a Blood paper with X. Jiang, I.Lossos and colleagues (2010, PubMed 20844236) - see panel E and F of https://pubmed.ncbi.nlm.nih.gov/20844236/#&gid=article-figures&pid=figure-2-uid-1 for HeLa "tetrads" of 4 grand-daughters 40 hours after plating dilute suspension; fit very nicely in 63x objective lens 2x confocal microscope zoom -- helps that HeL are not particularly motile; we also did 3-plane stacks, 0um, 2um and 4um from the coverglass-media surface, and reflection interference contrast microscopy to foucs consistenttly at the coverglass-media interface). If more cells are needed, users could optimize trypsinization, cell density, culture time, for their needs (ex: 60 hours would probably result in 8-cell HeLa colonies). Optimizing cell density at plating would also provide good density of colonies, not too sparse (ineffieinct searching) and not too dense overlap). * only two slides imaged ... you and your P.I. decide on what controls to do. If weird stuff appears in your images, if you run the appropriate controls every time, you can troubleshoot (provided you use 60x oil Z-series deconvolution when looking for subcellular details). If you do not have the controls, your choice, your thinking. ---------- Unknown: would there have been more information -- compared to the 60x/1.35NA lens -- if a) FISHscope has a 100x/1.4 NA objective lens (~60 nm pixel size) or 150x/1.45 NA (43 nm pixel size)? b) FISHscope wdetecting direct labeled antibody using BV421 (emission peak ~420nm) or BUV395 (emission peak ~395nm) instead of Alexa Fluor 488 (emission peak ~520nm)? c) confocal with the same lenses (i.e. move specimen to our Olympus FV3000RS confocal with same lenses)? Note: 108nm is FISHscope pixel size with the 60x/1.35NA objective lens using our Hamamatsu ORCA-FLASH4.0LT with 6.5x6.5um pixels (please donate to us the Hamamatsu Quest2 back-illuminated quantitative CMOS camera, 4.6x4.6um pixel size, peak 95% quantum efficiency s peak ~85% for 4.0LT) dxy is the XY resolution, My rule of thumb is dxy / 3 = optimum pixel size (though dxy / 3.5 might provide more information) -- basically Nyquist sampling theorem (2.2 data points for a sine wave) for 2D biological specimens. I also note deconvolution improves resolution by ~10% (if specimen, coverglass #1.5H, bright labeling, excellent camera, excellent optics and calculations are perfect, and no photobleaching). To make absolutely clear: We prioritized 60x/1.35 NA objective lens for FISHscope for single molecule RNA FISH: 108x108nm pixel size results in more area (~11,664 um^2) compared to 100x lens (65x65um pixels = 4225 um^2, 36%(calculations assume 100% fill factor for the pixels - probably not the case). For smFISH, pohotons matter more than resolution. dxy widefield = 0.61 * wavelength / NA dxy confocal 1.0 Airy unit) = 0.51 * wavelength / NA dxy confocal 0.66 Airy units = 0.8 * 0.51 * wavelength / NA ... 0.51 * 0.8 = 0.41. dxy (WF) = 0.61 * 500 nm / 1.35 = 226nm ... deconv (0.9) = 203nm ... 108nm pixel size too big on purpose. dxy (WF) = 0.61 * 500 nm / 1.40 = 217nm ... deconv (0.9) = 196nm ... ~60nm pixel size good match (3x is 180nm). dxy (WF) = 0.61 * 500 nm / 1.45 = 210nm ... deconv (0.9) = 189 nm ... 189/3.5 = 43nm oixel size, maybe more info. skipping 1.0 Airy unit and just calculating 0.66 Airy units ("confocal sweet spot" re Jeff Reece, NIH, sweet range). dxy (0.66AU) = 0.41 * 500 nm / 1.35 = 152nm ... deconv (0.9) = 137nm dxy (0.66AU) = 0.41 * 500 nm / 1.40 = 146nm ... deconv (0.9) = 132nm dxy (0.66AU) = 0.41 * 500 nm / 1.45 = 141nm ... deconv (0.9) = 127nm what if use wavelength 395nm (BUV395, ignoring light source issue wrt confocal): dxy (WF) = 0.61 * 395 nm / 1.45 = 166nm ... deconv (0.9) = 150nm dxy (0.66AU) = 0.41 * 395 nm / 1.45 = 111nm ... deconv (0.9) = 101nm I note that the last value, 101nm is better than commercial SIM (structured illumination microscope) systems, which is ~2x better resolution than standard 10 Airy unit confocal (but Alexa Fluor 488 etc -- SIM systems may or not be capable of exvitation of BUV395 and would not have BUV filter set standard). |
|
20241014M Coverglass info: No. 00 ~70 um (0.07 mm) No. 0 ~100 um (0.08 - 0.12mm) No. 1.5H 170 +/- 5 um (High precision) Ex: Marienfeld (Germany). notes: * I strongly recommend buying and routinely using #1.5H coverglass * #00 ... I communicated with Marienfeld (10/2024) ... #00 orders are doable, minimum 1000 pieces ... in the U.S., order through one of their two U.S. distributors (see their distributor wb page for contact info). * Sometimes 2 coverglasses can be stuck together when you take them out of the box. I encourage checking EVERY coverglass for "pairs". A pair can be recognized by Newton's Rings (interference fringes, similar to a thin layer of oil on a water puddle) appearing when you hold the coverglass(es) up to a light (ceiling light works fine). Positive control: you can hold 2 together. Another method is inspecting the edge: 2 stuck together appears thicker. You can also inspect the coverglass on edge (10x objective lens has a 1+ mm field of view; or use a mono-zoom or stereo microscope). * wrong coverglass thickness can result in spherical aberrations. * GM: if you refractive index match the immersion media (i.e. oil), coverglass (R.I. 1.518), and mounting media (i.e. cured Prolong Glass, R.I. 1.518) the light rays travel in a straight line = best imaging quality. This implies you could use a #00 or #0 or #1 coverglass and get additional working distance to see deeper into your specimen. |
|
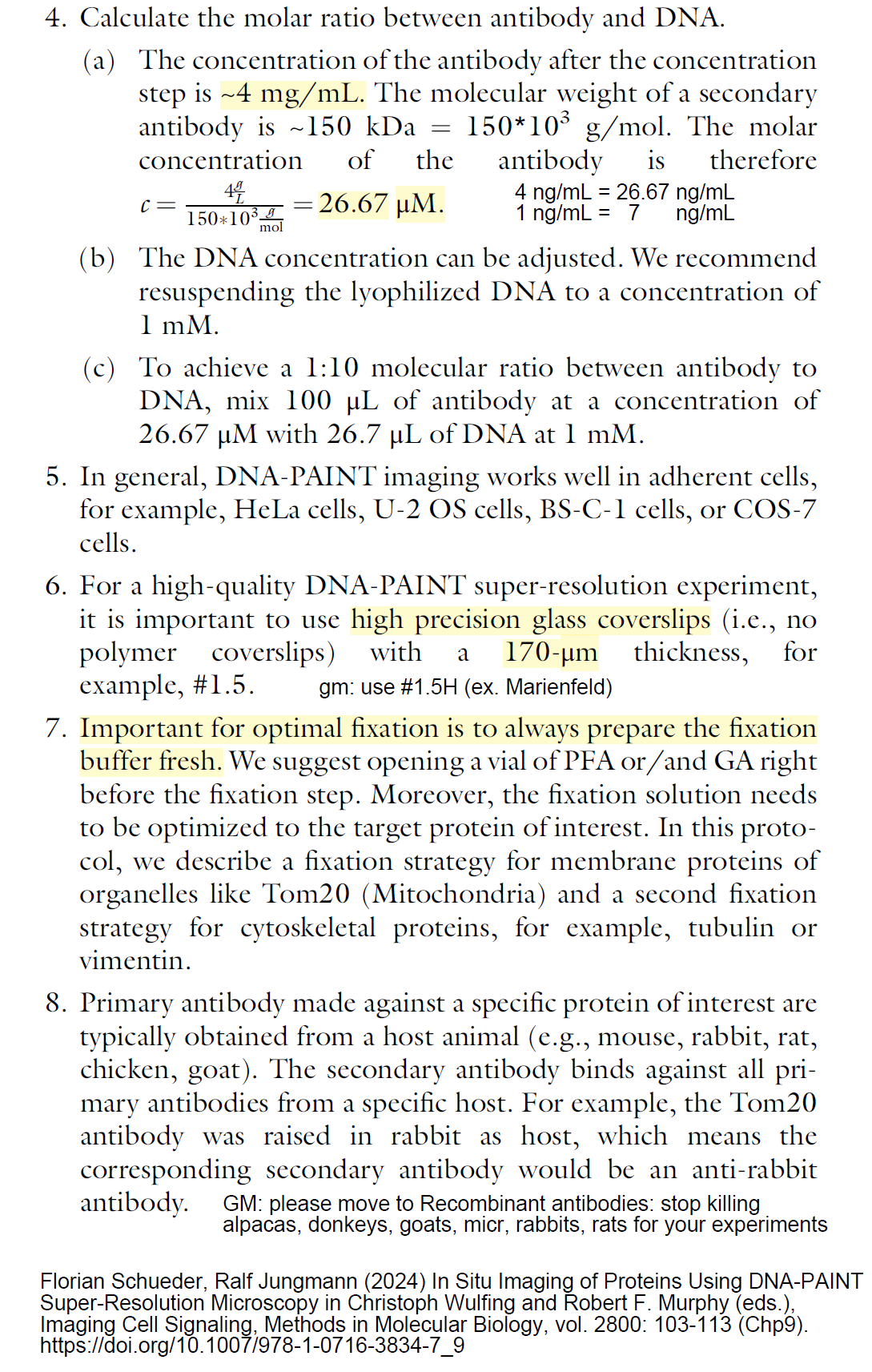
20241014M Amount of antibody in a typical immunofluorescence experiment or other antibody experiment page 111 (pdf page 11): Typical antibody is 100 ug in 100 uL, so 1 ug/ul = 1 mg/mL. I note that a Nanobody (VHH-only heavy chain, usually from alpaca or llama, aka "camelids") is ~15 kDa, so all concentrations would be 10x higher. See Nano-Tag.com and Chromotek - now https://www.ptglab.com/ (in U.S. distributed through ThermoFisher) for commercial fluorescent dye labeled secondary antibodies (both sourced from pleiner et al 2018 J Cell Biol). Florian Schueder, Ralf Jungmann (2024) In Situ Imaging of Proteins Using DNA-PAINT Super-Resolution Microscopy in Christoph Wulfing and Robert F. Murphy (eds.), Imaging Cell Signaling, Methods in Molecular Biology, vol. 2800: 103-113 (Chp9). https://doi.org/10.1007/978-1-0716-3834-7_9
|
|
20241011F GM favorite tools (mostly software) X1.com ... Windows PC indexing program. NCH Debut ... screen and audio recording software (see OBS Studio to have multiple webcams on screen). Drive cloning softweare - examples
SSD drives: NVMe M.2 drives 1TB (TeraByte) capacity and largers are usually (PCIe gen3, x4) ~3 GB/sec (3 GigaByte/sec ~ 3000 MegaByte/sec) vs 256 GB and 512 GB NVMe typically only 600 GB/sec, same as mSATA and SATA-6 SSDs. Motherboard performance (x16 slot) ... so NVMe drive x4 is 1/4 speed for x16 slot - can get Add-In Card (AIC) that can house 4 (or more) NVMe drives for one x16 slot (see RAID info below):
Resizable BAR = Modern (circa 2020 plus - usually later gen3 and newer genX) motherboards feature to turn on.
GPU (NVidia GPU cards dominates scientific software such as spatial deconvolution are mostly written only for NVidia CUDA driver interface) (Graphical RAM speed typically increase per generation): TITAN (circa 2023): ~3Teraflop (FP32) (probably PCIe first gen) RTX 2080 (circa 2016); "few Teraflops" (10?) PCIe gen2? RTX 3090 (circa 2019); ~20 Teraflops ... PCIe x16 gen3 RTX 4090 (circa 2022); ~90 Teraflops ... PCIe x16 gen4 RTX 5090 (expected Feb 2025); ~120 Teraflops ... PCIe x16 gen5 GM suggestion for NVidia and AMD: since top ofthe line GPUs are 2 or 3 PCIe slots wide to accomodate chillers and fans, add second PCIe x16 gen5 interface to DOUBLE or TRIPLE the data transfer rate. Sure, more complexity, so what? NVMe drives and arrays ... current Highpoint RAID PCIe gen5 AIC (add-in card) ~61 GigaBytes / second. Example:
*** [this sub-section added 20241020S[ RAM speeds - faster is better, and more expensive ... and need newer motherboard etc Current (2024) DDR6 PC ram speeds in Gigabytes/sec - for comparison with PCIe x16 slot speeds, NVMe drive arrays, GPU transfer speeds (and resizable BAR memory transfer aperture)
*** Hamrick.com VueScan Pro - universal scanning software for flatbed scanners and some 35mm film scanners. Flatbed scanner(s) - ideally with transparency lid. 35 mm film scanner(s) - with adapter(s) for microscope slides, maximum area is 36x24mm, examples:
CrystalDiskMark 8 and CrystalDiskInfo (8) ... a bit tricky finding the "download installer" links (buttons) on the web site
OBS Studio ... enables simultaneous use of multiple web cameras ... GM recommends using different model for each web cam to help keep organized. Glary Disk Cleaner ... weekly purge junk files from Windows PCs - avoid installing "Accessories" (junk). Gary Undelete ... best to install before you need to recover any files! - avoid installing "Accessories" (junk). PC speakers - I now recommend buy with AC power, generally enables louder output than USB powered. For work, no woofer (bass); for home but one with woofer (bass). Uni-Ball 0.7mm black pens; red pens. Sharpies - "magic markers" - available in many colors and formats. Tools (literally):
|
|
Yes, 63x /1.4NA objective lens will typically be more useful than a 100x/1.4 NA objective lens, for the larger field of view advantage you mentioned. This ASSUMES they are each in “perfect” condition. If either has been old and abused, the better quality one will be superior.
Confocal resolution … then AiryScan (divide confocal 1.0AU by 2) then pixel size (divide AiryScan in this example by 3.5:
Dxy (1.0 Airy Unit)= 0.51 * Lambda / NA Alexa Fluor 488 would then be 0.51 * 520nm / 1.4 = 189nm … so AiryScan divide by 2 = 90nm … pixel size 25.7nm … I would round down to 25nm. Brilliant Violet 421 0.51 * 440nm / 1.4 = 160nm … so AiryScan divide by 2 = 80nm … pixel size 22 nm … I would round down to 20nm.
Z-step size: the Z-resolution equation is complicated enough for me to avoid memorizing it, so I simplify that for high NA objective lenses, and skipping some steps, I would use
Z-step(AiryScan) as 3x the XY pixel size, so: Alexa Fluor 488 … 75nm. Brilliant Violet BV421 … 60nm.
Note: the best resolution for AiryScan requires doing the post-processing step(s) – I don’t know if this is “double deconvolution” or some other term. Also, do not assume Zeiss has the best math! You might want to get a demo license (2 weeks on one of your lab computers) of Huygens, see https://svi.nl/Huygens-Array-Detector-Software which may compute better.
**
If you are going to do AiryScan “at the limit’ a lot, you may want to contact our local Zeiss rep to get a demo of a new, higher NA, objective lens (and find the money to buy it if it works great). Zeiss offers 1.45NA objective lenses – usually for TIRF, but with perfect refractive index matching can acquire beautiful confocal or AiryScan, every 0.05 NA improvement increases resolution (if perfect specimen, coverglass etc). These lenses have limited working distance (say 100 um vs 200um for 1.40 NA lenses), but you want to image near the coverglass (under 20um) anyway.
àlastly, quick trick: if Zeiss has limited line averaging, can do line and frame averaging … can also do smaller Z-step size (say 2x as many) which the deconvolution software will more or less treat as equivalent to extra averaging.
|